1. Introduction
Human Pluripotent stem cells (hPSCs), including hESCs that first isolated by Thomson et al. [1] in 1998 and hiPSCs established in 2006–2007 by Yamanaka’s group [2,3], retain the core characteristics of self‑renewal and pluripotency, the capacity to differentiate into all three germ layers. So that, these unique properties have established hPSCs as a versatile platform for studying disease modeling, drug screening and regenerative medicine.
Early hPSC differentiation strategies, such as embryoid body formation and feeder-layer co-culture with mitotically inactivated mouse embryonic fibroblasts (MEFs), provided proof of concept for in vitro lineage specification by supplying extracellular matrix components and soluble inductive cues [1,4]. At that time, MEF feeders were essential for maintaining hESC pluripotency due to the absence of fully defined culture systems. However, the development of feeder-free, chemically defined media—such as mTeSR1 [5] and E8 medium [6]—combined with human-derived extracellular matrix coatings (e.g., vitronectin, laminin-521) has eliminated the need for mouse-derived feeder cells, enabling xeno-free culture conditions suitable for clinical-grade cell manufacturing. These advances, together with the identification of lineage-inducing factors—key transcription factors (e.g., SOX17, GATA4), growth factors, and small molecules like CHIR99021 —have greatly improved differentiation efficiency and reproducibility.
Because hPSCs have the inherent ability for unlimited self-renewal, theoretically they can be infinitely expanded and continuously differentiate into the target cells. Therefore, various cell types derived from hPSC (human pluripotent stem cells) can be produced on a large scale. Traditional drug screening often relies on animal models or limited human samples. However, hPSCs can continuously provide human-derived functional cells (such as neurons and cardiomyocytes), which are closer to the human physiological state. Such characteristics make them suitable for use in high-throughput drug screening, and hPSCs (especially iPSCs from patients themselves) can also be used for the development of personalized cell therapy [7].
Nevertheless, there are still challenges, including suboptimal differentiation efficiency, cellular heterogeneity, and incomplete functional maturation in vivo. In addition, for hESCs, unresolved ethical concerns surrounding embryo use continue to represent a major barrier to their clinical application.
Here, this paper overviews the most recent progress in understanding the underlying mechanisms of hPSC differentiation with emphasis on gene-editing technologies for directed differentiation, using small molecules as modulators, and 3D culture systems.
2. Gene editing technologies in hPSC differentiation
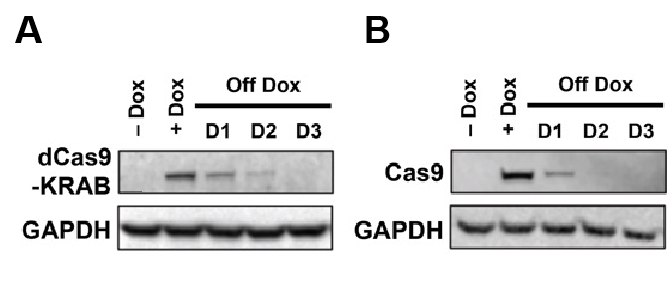
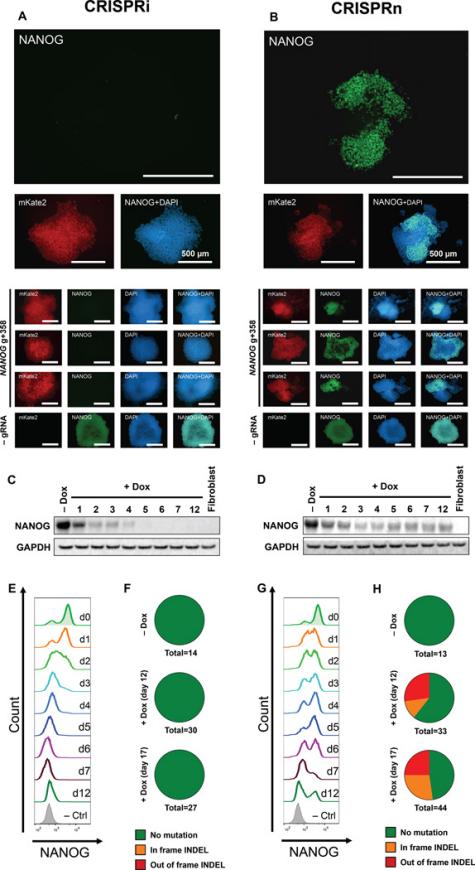
CRISPR/Cas9 technology has transformed the study of hPSCs by providing a versatile tool for precise and efficient genome editing. González et al. first established an inducible CRISPR (iCRISPR) platform in hESCs and hiPSCs, demonstrating robust and multiplexable gene editing [8]. Then inducible CRISPR interference and activation systems (CRISPRi/a) were developed, which enabled reversible and fine-tuned gene regulation in hPSCs [9]. Mandegar et al. (2016) proved that CRISPRi targeting OCT4 efficiently reduced its expression by more than 90%, making to loss of pluripotency and spontaneous differentiation, while CRISPRa targeting NANOG upregulated its transcription and then enhancing stem cell identity. Importantly, both effects were reversible upon doxycycline withdrawal, highlighting the potential of CRISPRi/a for finding gene function in hPSCs. As added in Figure 1 (A&B) and Figure 2, Mandegar et al. established doxycycline-inducible CRISPRi/a systems in hPSCs, presenting reversible regulation of Cas9 expression and functional control of pluripotency genes.
Subsequently, large-scale CRISPR-based functional genomic screens, often integrated with single-cell transcriptomics, have allowed systematic dissection of lineage regulators and stress response pathways. For example, Adamson et al. developed a multiplexed single-cell CRISPR screening platform that enabled high-resolution mapping of the unfolded protein response (UPR), providing a paradigm for how CRISPR perturbations can be systematically linked to transcriptional states at single-cell resolution [10].
Together, these methodological advances illustrate a trajectory from basic gene targeting towards scalable and high-resolution functional genomics, laying the foundation for rational optimization of hPSC differentiation protocols.
3. Small-molecule regulation of hPSC differentiation
Small molecules offer a powerful, cost-effective, and scalable means to direct hPSC differentiation with high reproducibility. Unlike growth factors, which suffer from high cost and batch variability, small molecules precisely modulate intracellular signaling pathways. For example, CHIR99021, a GSK3β inhibitor, robustly activates Wnt/β-catenin signaling to promote mesoderm induction, whereas SB431542 blocks Activin/Nodal signaling to prevent endodermal or ectodermal diversion.
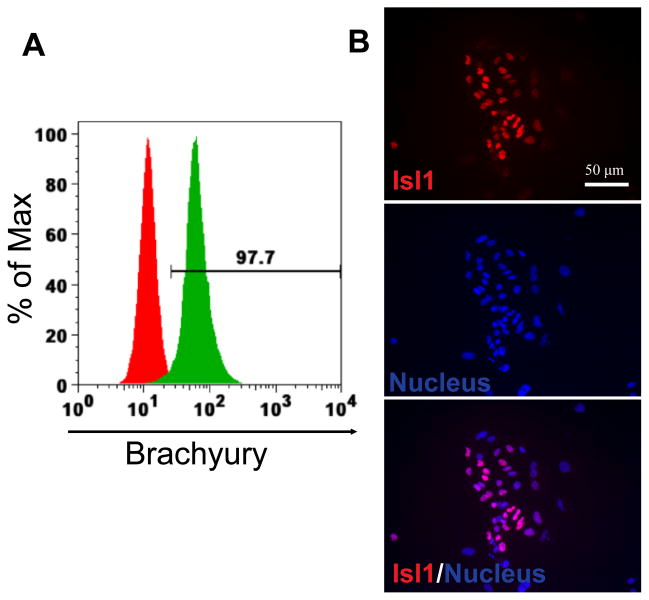
A typical example of small-molecule regulation is the cardiac differentiation protocol reported by Lian et al., which modulates Wnt signaling in a stepwise manner using the GSK3β inhibitor CHIR99021 followed by the Wnt inhibitor IWP2 under chemically defined conditions [11]. After 24 h of CHIR99021 treatment, ~98% of hESCs became Brachyury-positive, indicating highly efficient mesodermal induction (Figure 3A). By day 6, cells expressed the cardiac progenitor marker Isl1, confirming differentiation toward the cardiac mesoderm (Figure 3B).
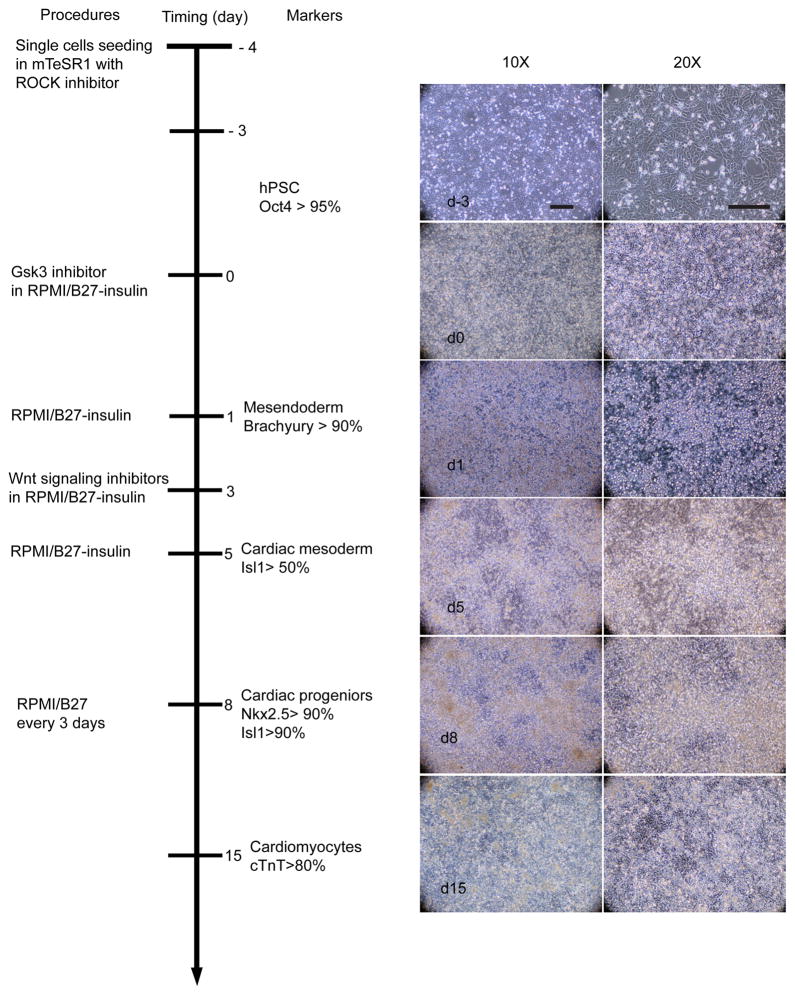
The overall time course of differentiation is summarized in Figure 4: hPSCs (Oct4+) transition into mesoderm (Brachyury+), then into cardiac mesoderm (Isl1+), and subsequently cardiac progenitors (Nkx2.5+/Isl1+), finally leading to >80% cTnT+ cardiomyocytes in day 15.
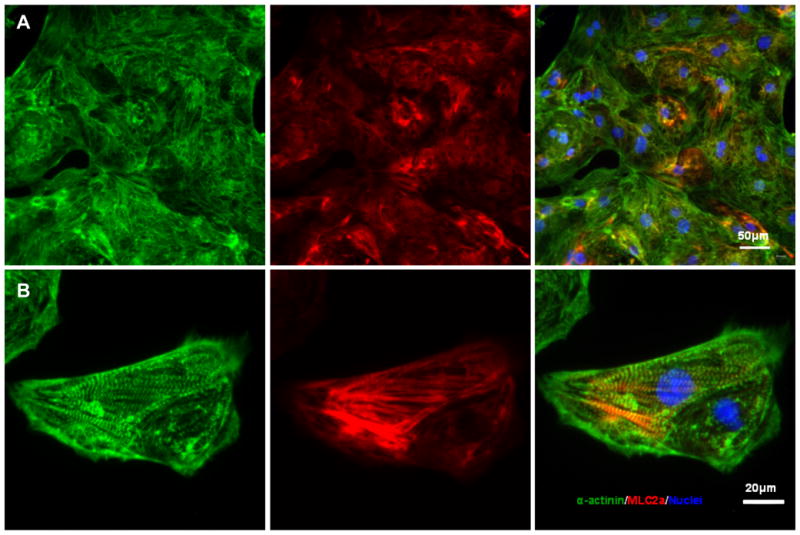
The study shows that, structural analysis of day 20 hPSC-derived cardiomyocytes revealed well-organized sarcomeric evidenced by immunostaining for α-actinin and MLC2a (Figure 5), proofing not only the efficiency but also the functional maturation of the differentiated cells.
Overall, this example highlights that staged small-molecule modulation of Wnt signaling provides a simple, reproducible, and growth factor–free strategy for directing hPSC differentiation into cardiomyocytes.
Similarly, Ma et al. reviewed that CHIR99021 combined with SB431542 significantly enhances lineage-specific outcomes across multiple protocols [12]. To sum up, small-molecule approaches enable more defined, reproducible, and clinically adaptable differentiation strategies.
4. 3D systems for enhancing hPSC differentiation
Three-dimensional (3D) culture systems better recapitulate the in vivo microenvironment compared to traditional two-dimensional monolayer cultures. By providing spatial organization, extracellular matrix interactions, and physiological gradients, 3D models improve differentiation efficiency and functional maturation.
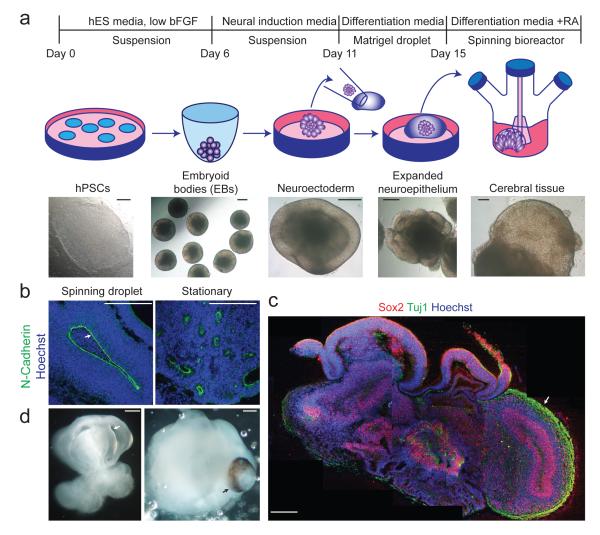
For example, Lancaster et al. established cerebral organoids from hPSCs that mimic human cortical development and enable disease modeling of microcephaly [13].
They developed a cerebral organoid culture system by embedding hPSCs in Matrigel droplets and maintaining them in spinning bioreactors, which provided sufficient nutrient and oxygen exchange to support long-term growth. Brain tissues developed by this way showed a very high efficiency, which needs only 8-10 days to make the appearance of neural identity and form defined brain regions in only 20-30 days(Figure 6) .Within the cortical regions, radial glia, neural progenitors, and layered cortical neurons were observed, recapitulating key features of early human brain development. Importantly, when patient-derived iPSCs carrying CDK5RAP2 mutations were used, the resulting organoids were significantly smaller because of damaged progenitor proliferation, thus modeling the pathological features of microcephaly. This work established cerebral organoids as a powerful 3D platform for studying human neurodevelopment and neurodevelopmental disorders.
Similarly, Zhang et al. developed cardiac organoids with tissue-level organization using hPSC-derived cardiomyocytes and endothelial cells on 3D bioprinted scaffolds [14]. Building on these advances, Ronaldson-Bouchard et al. further demonstrated that applying mechanical and electrical stimulation to hPSC-derived cardiac tissues promotes advanced maturation, yielding structural, electrophysiological, and metabolic properties closer to those of adult human myocardium [15]. These 3D platforms provide not only mechanistic insights into human development but also translational models for drug testing and regenerative medicine.
5. Discussion
In recent years, gene editing technology, small molecule regulatory strategies, and three-dimensional culture system technologies have each provided different but complementary perspectives. The CRISPR-based gene editing platform enables researchers to truly pose causal questions: What role does a certain transcription factor, epigenetic regulatory factor, or signaling pathway play in the differentiation process, rather than merely making correlation observations? The small molecule regulation methods have fully demonstrated that precise regulation of developmental signals not only can replace expensive exogenous growth factors but also can improve the stability and reproducibility of experiments. The three-dimensional model also indicates that the tissues derived from hPSCs have the potential for self-organization and can form the same spatial structure as in the body, to a certain extent, they can also reproduce the key features of human development and even have certain advantages over traditional animal models in disease modeling. However, there are still some limitations. The differentiated cells generated by hPSCs are usually closer to the fetal state rather than mature adult cells, thus limiting their direct application in regenerative medicine. Although CRISPR technology has great potential, it still has safety issues, such as off-target effects and genomic stability, especially when considering clinical translation, more caution is needed. Although organoids have significantly provided a window for studying human development, they still lack the support of other systems. Therefore, although the hPSC differentiation research has made significant progress, it is still quite far from "completely solving" the problem.
In the future, the combination of these three cutting-edge technologies will not only facilitate a deeper understanding of human also lay the foundation for the clinical application prospects of pluripotent stem cells.
6. Conclusion
Human pluripotent stem cells (hPSCs), which include both human embryonic stem cells (hESCs) and human induced pluripotent stem cells (hiPSCs), have the capacity for infinite self-renewal and differentiation into virtually any tissue of the body, defining an unparalleled platform for regenerative medicine, disease modeling, as well as high-throughput screening.
In the last few years, significant progress has been reached in unraveling the molecular principles of hPSC differentiation and in optimizing culture systems to increase efficiency and standardization. These include recent developments in CRISPR/Cas9-mediated genome engineering, small-molecule–based lineage control, and three-dimensional differentiation platforms to direct hPSC fate at higher resolution and with increased physiological relevance.
However, there are challenges remaining in need of resolution, such as lack of full functional maturation of derived cells, cellular contamination by hESCs, noted genotype variability, and issues surrounding the ethics of using hESCs. To this end, the future will have to overcome these limitations by integrating multi-omics approaches, genome editing, as well as recent bioengineering innovations.
In conclusion, improvement in hPSC differentiation, such as chemically defined media, will be essential for translating stem cell-based therapies to the clinic, opening a new era of personalized regenerative medicine.
References
[1]. Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998 Nov 6; 282(5391): 1145-7. doi: 10.1126/science.282.5391.1145. Erratum in: Science 1998 Dec 4; 282(5395): 1827. PMID: 9804556.
[2]. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006 Aug 25; 126(4): 663-76. doi: 10.1016/j.cell.2006.07.024. Epub 2006 Aug 10. PMID: 16904174.
[3]. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007 Nov 30; 131(5): 861-72. doi: 10.1016/j.cell.2007.11.019. PMID: 18035408.
[4]. Itskovitz-Eldor J, Schuldiner M, Karsenti D, Eden A, Yanuka O, Amit M, Soreq H, Benvenisty N. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Mol Med. 2000 Feb; 6(2): 88-95. PMID: 10859025; PMCID: PMC1949933.
[5]. Ludwig TE, Levenstein ME, Jones JM, Berggren WT, Mitchen ER, Frane JL, Crandall LJ, Daigh CA, Conard KR, Piekarczyk MS, Llanas RA, Thomson JA. Derivation of human embryonic stem cells in defined conditions. Nat Biotechnol. 2006 Feb; 24(2): 185-7. doi: 10.1038/nbt1177. PMID: 16388305.
[6]. Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, Propson NE, Wagner R, Lee GO, Antosiewicz-Bourget J, Teng JM, Thomson JA. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011 May; 8(5): 424-9. doi: 10.1038/nmeth.1593. Epub 2011 Apr 10. PMID: 21478862; PMCID: PMC3084903.
[7]. Shi, Y., Inoue, H., Wu, J. C., & Yamanaka, S. (2017). Induced pluripotent stem cell technology: a decade of progress. Nature reviews. Drug discovery, 16(2), 115–130. https: //doi.org/10.1038/nrd.2016.245
[8]. González F, Zhu Z, Shi ZD, Lelli K, Verma N, Li QV, Huangfu D. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014 Aug 7; 15(2): 215-226. doi: 10.1016/j.stem.2014.05.018. Epub 2014 Jun 12. PMID: 24931489; PMCID: PMC4127112.
[9]. Mandegar MA, Huebsch N, Frolov EB, Shin E, Truong A, Olvera MP, Chan AH, Miyaoka Y, Holmes K, Spencer CI, Judge LM, Gordon DE, Eskildsen TV, Villalta JE, Horlbeck MA, Gilbert LA, Krogan NJ, Sheikh SP, Weissman JS, Qi LS, So PL, Conklin BR. CRISPR Interference Efficiently Induces Specific and Reversible Gene Silencing in Human iPSCs. Cell Stem Cell. 2016 Apr 7; 18(4): 541-53. doi: 10.1016/j.stem.2016.01.022. Epub 2016 Mar 10. PMID: 26971820; PMCID: PMC4830697.
[10]. Adamson B, Norman TM, Jost M, Cho MY, Nuñez JK, Chen Y, Villalta JE, Gilbert LA, Horlbeck MA, Hein MY, Pak RA, Gray AN, Gross CA, Dixit A, Parnas O, Regev A, Weissman JS. A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell. 2016 Dec 15; 167(7): 1867-1882.e21. doi: 10.1016/j.cell.2016.11.048. PMID: 27984733; PMCID: PMC5315571.
[11]. Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ, Palecek SP. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat Protoc. 2013 Jan; 8(1): 162-75. doi: 10.1038/nprot.2012.150. Epub 2012 Dec 20. PMID: 23257984; PMCID: PMC3612968.
[12]. Ma X, Kong L, Zhu S. Reprogramming cell fates by small molecules. Protein Cell. 2017 May; 8(5): 328-348. doi: 10.1007/s13238-016-0362-6. Epub 2017 Feb 17. PMID: 28213718; PMCID: PMC5413596.
[13]. Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013 Sep 19; 501(7467): 373-9. doi: 10.1038/nature12517. Epub 2013 Aug 28. PMID: 23995685; PMCID: PMC3817409.
[14]. Zhang YS, Arneri A, Bersini S, Shin SR, Zhu K, Goli-Malekabadi Z, Aleman J, Colosi C, Busignani F, Dell'Erba V, Bishop C, Shupe T, Demarchi D, Moretti M, Rasponi M, Dokmeci MR, Atala A, Khademhosseini A. Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials. 2016 Dec; 110: 45-59. doi: 10.1016/j.biomaterials.2016.09.003. Epub 2016 Sep 5. Erratum in: Biomaterials. 2025 Nov; 322: 123363. doi: 10.1016/j.biomaterials.2025.123363. PMID: 27710832; PMCID: PMC5198581.
[15]. Ronaldson-Bouchard K, Ma SP, Yeager K, Chen T, Song L, Sirabella D, Morikawa K, Teles D, Yazawa M, Vunjak-Novakovic G. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature. 2018 Apr; 556(7700): 239-243. doi: 10.1038/s41586-018-0016-3. Epub 2018 Apr 4. Erratum in: Nature. 2019 Aug; 572(7769): E16-E17. doi: 10.1038/s41586-019-1415-9. PMID: 29618819; PMCID: PMC5895513.
Cite this article
Liu,W. (2025). Advances in Technologies Guiding Human Pluripotent Stem Cell Differentiation. Theoretical and Natural Science,141,101-108.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of ICBioMed 2025 Symposium: AI for Healthcare: Advanced Medical Data Analytics and Smart Rehabilitation
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998 Nov 6; 282(5391): 1145-7. doi: 10.1126/science.282.5391.1145. Erratum in: Science 1998 Dec 4; 282(5395): 1827. PMID: 9804556.
[2]. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006 Aug 25; 126(4): 663-76. doi: 10.1016/j.cell.2006.07.024. Epub 2006 Aug 10. PMID: 16904174.
[3]. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007 Nov 30; 131(5): 861-72. doi: 10.1016/j.cell.2007.11.019. PMID: 18035408.
[4]. Itskovitz-Eldor J, Schuldiner M, Karsenti D, Eden A, Yanuka O, Amit M, Soreq H, Benvenisty N. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Mol Med. 2000 Feb; 6(2): 88-95. PMID: 10859025; PMCID: PMC1949933.
[5]. Ludwig TE, Levenstein ME, Jones JM, Berggren WT, Mitchen ER, Frane JL, Crandall LJ, Daigh CA, Conard KR, Piekarczyk MS, Llanas RA, Thomson JA. Derivation of human embryonic stem cells in defined conditions. Nat Biotechnol. 2006 Feb; 24(2): 185-7. doi: 10.1038/nbt1177. PMID: 16388305.
[6]. Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, Propson NE, Wagner R, Lee GO, Antosiewicz-Bourget J, Teng JM, Thomson JA. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011 May; 8(5): 424-9. doi: 10.1038/nmeth.1593. Epub 2011 Apr 10. PMID: 21478862; PMCID: PMC3084903.
[7]. Shi, Y., Inoue, H., Wu, J. C., & Yamanaka, S. (2017). Induced pluripotent stem cell technology: a decade of progress. Nature reviews. Drug discovery, 16(2), 115–130. https: //doi.org/10.1038/nrd.2016.245
[8]. González F, Zhu Z, Shi ZD, Lelli K, Verma N, Li QV, Huangfu D. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014 Aug 7; 15(2): 215-226. doi: 10.1016/j.stem.2014.05.018. Epub 2014 Jun 12. PMID: 24931489; PMCID: PMC4127112.
[9]. Mandegar MA, Huebsch N, Frolov EB, Shin E, Truong A, Olvera MP, Chan AH, Miyaoka Y, Holmes K, Spencer CI, Judge LM, Gordon DE, Eskildsen TV, Villalta JE, Horlbeck MA, Gilbert LA, Krogan NJ, Sheikh SP, Weissman JS, Qi LS, So PL, Conklin BR. CRISPR Interference Efficiently Induces Specific and Reversible Gene Silencing in Human iPSCs. Cell Stem Cell. 2016 Apr 7; 18(4): 541-53. doi: 10.1016/j.stem.2016.01.022. Epub 2016 Mar 10. PMID: 26971820; PMCID: PMC4830697.
[10]. Adamson B, Norman TM, Jost M, Cho MY, Nuñez JK, Chen Y, Villalta JE, Gilbert LA, Horlbeck MA, Hein MY, Pak RA, Gray AN, Gross CA, Dixit A, Parnas O, Regev A, Weissman JS. A Multiplexed Single-Cell CRISPR Screening Platform Enables Systematic Dissection of the Unfolded Protein Response. Cell. 2016 Dec 15; 167(7): 1867-1882.e21. doi: 10.1016/j.cell.2016.11.048. PMID: 27984733; PMCID: PMC5315571.
[11]. Lian X, Zhang J, Azarin SM, Zhu K, Hazeltine LB, Bao X, Hsiao C, Kamp TJ, Palecek SP. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/β-catenin signaling under fully defined conditions. Nat Protoc. 2013 Jan; 8(1): 162-75. doi: 10.1038/nprot.2012.150. Epub 2012 Dec 20. PMID: 23257984; PMCID: PMC3612968.
[12]. Ma X, Kong L, Zhu S. Reprogramming cell fates by small molecules. Protein Cell. 2017 May; 8(5): 328-348. doi: 10.1007/s13238-016-0362-6. Epub 2017 Feb 17. PMID: 28213718; PMCID: PMC5413596.
[13]. Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013 Sep 19; 501(7467): 373-9. doi: 10.1038/nature12517. Epub 2013 Aug 28. PMID: 23995685; PMCID: PMC3817409.
[14]. Zhang YS, Arneri A, Bersini S, Shin SR, Zhu K, Goli-Malekabadi Z, Aleman J, Colosi C, Busignani F, Dell'Erba V, Bishop C, Shupe T, Demarchi D, Moretti M, Rasponi M, Dokmeci MR, Atala A, Khademhosseini A. Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials. 2016 Dec; 110: 45-59. doi: 10.1016/j.biomaterials.2016.09.003. Epub 2016 Sep 5. Erratum in: Biomaterials. 2025 Nov; 322: 123363. doi: 10.1016/j.biomaterials.2025.123363. PMID: 27710832; PMCID: PMC5198581.
[15]. Ronaldson-Bouchard K, Ma SP, Yeager K, Chen T, Song L, Sirabella D, Morikawa K, Teles D, Yazawa M, Vunjak-Novakovic G. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature. 2018 Apr; 556(7700): 239-243. doi: 10.1038/s41586-018-0016-3. Epub 2018 Apr 4. Erratum in: Nature. 2019 Aug; 572(7769): E16-E17. doi: 10.1038/s41586-019-1415-9. PMID: 29618819; PMCID: PMC5895513.









