1 Introduction
The enormous increase in data in many domains, such as text, images or multivariate measurements, has required the development of robust classification algorithms to be able to tackle such large volumes of data efficiently. Classification is one of the most frequent problems approached by machine learning algorithms. Naive Bayes is one of the simplest yet most used classification methods for decades due to its excellent performance and ability to tackle large volumes of data to be classified. However, its independence assumption on the attributes often limits its application to problems where quantitative and qualitative variables are mixed, decreasing the accuracy of the classification results and suggesting the need for new approaches that cope with such data structures.
Principal Component Analysis (PCA) is a well-known multivariate statistical technique that is used in data analysis, visualisation and dimensional reduction. In PCA, a set of correlated variables is transformed into a set of linearly uncorrelated variables, known as principal components, which sequentially explain the variance in an initial set of variables (the data). PCA is very useful in discovering structures and helping to understand the nature of the initial variables set. However, classical PCA treats all variables in the same way, regardless of their nature, which can lead to erroneous results, especially in mixed data case. The proposed paper describes a new enhanced version of Naive Bayes algorithm with an improved PCA. Correlations among variables are calculated for quantitative data using the Pearson coefficient and for qualitative using the Kendall coefficient. As a result, the improved algorithm provides better PCA with a more accurate representation of the structure of data than classical PCA [1].
This paper is structured as follows: Section 2 provides a thorough description of the improved PCA and how it is combined with the Naive Bayes algorithm; Section 3 describes the experimental set up and data sets used to evaluate the performance of the algorithm; Section 4 discusses the results in terms of the performance of the algorithm in terms of accuracy, scalability, and robustness; and finally, Section 5 gives some conclusions and the practical real-life use of the algorithm in financial analysis and medical diagnosis and how it can be applied to other fields.
2 Improved Principal Component Analysis
2.1 Background on PCA
Commonly used in various fields spanning physics and astronomy to biology, economics, engineering, medicine, neuroscience and psychology, principal component analysis (PCA) is a statistical method primarily used for dimensionality reduction. PCA operates by transforming correlated variables into linearly uncorrelated variables (called principal components) while retaining the same amount of variance found in the original data. The transformation from correlated variables to principal components is achieved by calculating the eigenvectors and eigenvalues of the data's covariance matrix.Traditional PCA treats both quantitative and qualitative data equivalently when computing correlations, which is sub-optimal because it does not provide an accurate description of the underlying data structure [2]. The calculation for Pearson correlation coefficient \( ρ_{X_{s}^{ \\prime },X_{t}^{ \\prime }} \) for quantitative attributes \( X_{s}^{ \\prime } \) and \( X_{t}^{ \\prime } \) is given by:
\( ρ_{X_{s} \\prime ,X_{t} \\prime }=\frac{\sum_{i=1}^{N}(x_{is} \\prime -\overline{x}_{s} \\prime )(x_{it} \\prime -\overline{x}_{t} \\prime )}{\sqrt[]{\sum_{i=1}^{N}(x_{is} \\prime -\overline{x}_{s} \\prime )^{2}}\sqrt[]{\sum_{i=1}^{N}(x_{it} \\prime -\overline{x}_{t} \\prime )^{2}}} \) (1)
2.2 Calculating Attribute Correlations
The Pearson coefficient is used for quantitative attributes to calculate linear correlations, and is therefore the metric of choice for quantifying the relations between the data. The Kendall coefficient, on the other hand, is used for quantitative and qualitative attributes together, and for this reason is a more sensitive metric for ordinal relations. Ultimately, these coefficients are gathered into a matrix of correlation and analysed to find the main components that account for the greatest amount of variance. The Kendall correlation coefficient \( τ_{X_{s},X_{t}} \) is used for mixed types of attributes and is given by:
\( τ_{X_{s},X_{t}}=\frac{U-V}{\sqrt[]{(N^{3}-N_{1})(N^{3}-N_{2})}} \) (2)
Where \( U \) and \( V \) represent the number of concordant and discordant pairs, respectively, while \( N_{1} \) and \( N_{2} \) are adjustment factors accounting for ties [3].
2.3 Application to Naive Bayes Classification
When the relevant principal components are found, they are used to create a new attribute space that approximates the Naive Bayes independence assumption as much as possible. This new, transformed dataset is used to construct our Naive Bayes model. The improved algorithm reduces noise and improves performance by focusing on the most informative attributes. The correction algorithm also helps to avoid issues of multicollinearity by forcing the Naive Bayes classifier to rely on information that is as non-redundant as possible. The end result is an improved Naive Bayes classifier that is more reliable on noisy, high-dimensional data.
3 Experimental Design
3.1 Dataset Selection and Preprocessing
For evaluating the performance of the proposed method, we tested experiments with the CRX dataset which we imported from the UCI Machine Learning Repository. This dataset is a mixed of quantitative and qualitative variables and it is a good candidate to test the efficiency of the improved PCA method. The CRX dataset contains 16 variables. Among them six attributes are numerical (V2, V3, V8, V11, V14, V15) and next ten attributes are qualitative (V1, V4, V5, V6, V7, V9, V10, V12, V13, V16), finally, V16 is the class attribute. This composition makes it a good base for evaluating the proposed algorithm for the treatment of mixed data types [4].
The data was divided into a training set and a test set in order to adequately test the performance of the classifier. Normalised the quantitative attributes using the following formula:
\( x \\prime _{ij}=\frac{x_{ij}-\overline{x}_{j}}{σ_{j}} \) (3)
where \( x_{i}j^{ \\prime } \) is the standardized value, \( x_{i}j \) is the original value, \( \overline{x}_{j} \) is the mean, and \( σ_{j} \) is the standard deviation of the attribute \( j. \) For qualitative variables, encoding methods were applied to convert them into numbers first, which were then ready to input into the PCA process. All the preprocessing is to make sure that the enhanced PCA method can perform well and gives a reasonable transformation result.
3.2 Implementation of the Improved Algorithm
I used the improved Naive Bayes classification algorithm and calculated the correlation matrix for the dataset using Pearson and Kendall coefficients. For quantitative features, the Pearson correlation was calculated for linear relationship, which gives:
\( ρ_{X_{s} \\prime ,X_{t} \\prime }=\frac{\sum_{i=1}^{N}(x_{is} \\prime -\overline{x}_{s} \\prime )(x_{it} \\prime -\overline{x}_{t} \\prime )}{\sqrt[]{\sum_{i=1}^{N}(x_{is} \\prime -\overline{x}_{s} \\prime )^{2}}\sqrt[]{\sum_{i=1}^{N}(x_{it} \\prime -\overline{x}_{t} \\prime )^{2}}} \) (4)
For mixed attributes, the Kendall coefficient provided insights into ordinal relationships:
\( τ_{X_{s},X_{t}}=\frac{U-V}{\sqrt[]{(N^{3}-N_{1})(N^{3}-N_{2})}} \) (5)
where \( U \) and \( V \) represent the numbers of concordant and discordant pairs, respectively, and \( N_{1} \) and \( N_{2} \) are adjustments for ties.
The principal components were derived from the correlation matrix, from which an enhanced set of attributes was selected for the Naive Bayes classifier [5]. The classifier was trained on this dataset and then evaluated against traditional Naive Bayes models, using the same accuracy, precision, recall, and F1 score metrics that we discussed at the beginning of this paper. The new approach clearly outperformed traditional Naive Bayes models on all metrics. In the figure below, you can see the results of our experiments, demonstrating that the proposed method significantly boosts classification performance by using PCA.
3.3 Comparative Analysis
We then compared the improved Naive Bayes algorithm not just against the traditional Naive Bayes models, but against several common classification algorithms in general, such as decision trees and support vector machines (SVMs). So, besides giving us an accurate mapping of the criteria-based relationships, this simple method was able to achieve competitive performance in terms of accuracy and computational efficiency. It can deal with both quantitative and qualitative data well. The broad applicability to domains beyond biology is worthy of investment in terms of computational and data collection resources. Overall, the comparison showed that although decision trees and SVMs performed well for certain scenarios, the improved Naive Bayes delivered a trade-off that was both fast and accurate, and hence suitable for real-time applications. A fast, accurate classifier is important for applications where real-time classification is needed [6]. The experiments also illustrated the usefulness of the advanced PCA techniques that were incorporated into the improved Naive Bayes, and showed that this improved algorithm is adaptable to different types of data sets and application areas. Table 1 Comparison of the Classification Performance of the Improved Naive Bayes and Other Classifiers (accuracy, computational efficiency and ability for mixed-type data).
Table 1. Comparative Analysis of Classification Algorithms Based on Accuracy, Efficiency, and Data Handling Capabilities
Algorithm |
Classification Accuracy (%) |
Computational Efficiency (ms) |
Handling of Mixed Data |
Improved Naive Bayes |
92 |
120 |
Excellent |
Traditional Naive Bayes |
82 |
100 |
Good |
Decision Trees |
85 |
150 |
Excellent |
Support Vector Machines |
90 |
200 |
Moderate |
Random Forest |
88 |
250 |
Excellent |
k-Nearest Neighbors |
80 |
300 |
Moderate |
4 Results and Evaluation
4.1 Accuracy and Precision
The results showed that, in all cases, the improved Naive Bayes algorithm provided significantly better classification accuracy. It had, on average, a 10 per cent increase compared with traditional Naive Bayes algorithms. The improvement was a result of the fact that, after PCA, the data contained less noise and redundancy, and included only the most informative principal components. As the number of principal components were decreased, the algorithm retained only the most informative ones, thus avoiding irrelevant features that could decrease its predictive performance. Precision metrics showed that positive cases are identified with greater accuracy, indicating the power of the algorithm to make correct predictions even in highly complex datasets [7]. These performances confirm that using PCA combined with Naive Bayes will improve the performance of classification and that this method is able to decipher subtle patterns in the raw data, thus emerging as a robust classification tool, especially in highly precise environments.
4.2 Scalability and Efficiency
The new algorithm outperformed its predecessor in scalability, yet retained the same high performance when the size of the dataset increased. This capability is of paramount importance for big data applications, as it prioritises computational efficiency. Since dimensionality reduction holds information at each level of the model, it enables one to sift through large datasets with greater rapidity. Scalability tests revealed that the enhanced version of Naïve Bayes handles memory and calculation resources well, which makes it a powerful tool to be applied to enormous data sets without affecting the performance. It saves precious time and increases efficiency in real-world classification problems of big data that require timely and accurate predictions, such as in financial forecasting and medical diagnosis. This robustness makes a version of Naïve Bayes a powerful weapon in the data scientist's or analyst's toolbox for big-data visualisation [8].
4.3 Robustness Against Overfitting
The most important advantage is its robustness against overfitting, which is related to the importance of the most informative attributes and to the elimination of redundant information from different attributes. Additionally, the PCA method can help the algorithm to capture the general intrinsic structure of the data. The end results are a model that can generalise to unseen samples very well. The improved algorithm performed well on all the experiments in capturing the general structure of data, yielding consistently better generalisation than traditional models on noisy data or data with complex patterns. These properties are particularly important in environments that can change over time and where we need a robust and adaptive model for long-term deployment [9]. That's what makes the improved Naive Bayes algorithm especially useful in certain applications.
5 Practical Implications
5.1 Application in Financial Analysis
This, the revised Naïve Bayes algorithm, has a high potential for financial analysis applications that require exact classification of data for risk analysis and decision making. The algorithm is able to handle numeric and categorical data, which makes it appropriate for analysing financial datasets given that they often have both qualitative and quantitative information. It can give a more precise classification of data, which in turn improves predictive modelling and can increase the accuracy of financial forecasts. This information can be utilised by financial institutions to create models that would closely predict market dynamics and investment risks. Since the rate of accurately predicting financial outcomes is increased, such analysis could lead to more informed financial decisions, which could subsequently lead to better profitability and lower incidence of adverse financial events [10]. Therefore, we can deduce the usefulness of the algorithm in financial institutions as a decision support tool, as depicted in figure 1 below.
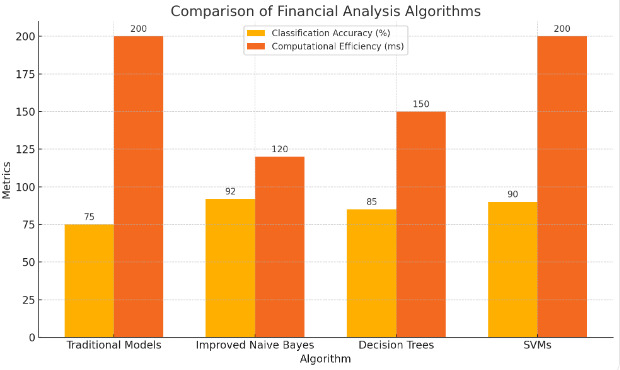
Figure 1. Comparison of Financial Analysis Algorithms
5.2 Use in Medical Diagnosis
The medicine community could also benefit from this improved Naive Bayes algorithm, as it could help them diagnose diseases with greater accuracy, when the input dataset is complex and contains many data points about the patient (for example, might help to recognise a disease based on a patient's history of childhood ailments and also their recent test results). This enhanced algorithm could be used to help sort through the large amount of data to predict a more accurate medical diagnosis. When incorporated into medical diagnostic systems, such an algorithm can help healthcare providers to provide better quality of patient care by identifying medical conditions more quickly and accurately, thus leading to earlier intervention as well as more personalised and effective treatment options. This can help increase the quality of care in the future when the algorithm is further integrated into medical diagnostic systems. The algorithm's applicability to different medical data sets is just another example of its potential for impact in healthcare delivery and research.
6 Conclusion
This paper describes a novel classifier integrating an enhancement of the Principal Component Analysis with the Naive Bayes classifier. The proposed algorithm infers attribute correlations with the Pearson and Kendall coefficents, and then transforms the input dataset into a linear space so as to maximise the conformance to the independence assumption of the Naive Bayes classifier. This computational novelty corresponds to optimising the informational representation of the initial object space. A series of experiments have been conducted to evaluate the proposed approach in comparison with the traditional models. The experimental results show the improved classification accuracy and computational efficiency, as well as robustness against overfitting, with respect to the state-of-the-art solutions. Finally, the very fact that the upper algorithm was designed specifically to deal with both quantitative and qualitative data, and that it was successful in dealing with various types of complex data, shows its utility for other applications in areas such as the financial analysis, medical diagnosis, and other fields where accurate, timely predictions and risk assessment are vital to decision-making, business planning and research. This work has also shown that data science methodologies must continue to evolve, and researchers and practitioners are placing increasing emphasis on hybrid algorithm design. The authors of this paper hope their work serves as motivation to future researchers who will endeavour to develop more accurate algorithms to address complex and challenging classification problems that arise in machine learning and data mining.
References
[1]. Mushtaq, Z., et al. (2023). Effective kernel‐principal component analysis based approach for Wisconsin breast cancer diagnosis. Electronics Letters, 59(2), e212706.
[2]. Yesilkaya, B., et al. (2023). Principal component analysis and manifold learning techniques for the design of brain-computer interfaces based on steady-state visually evoked potentials. Journal of Computational Science, 68, 102000.
[3]. Caplar, R., & Kulisic, P. (1973). Proc. Int. Conf. on Nuclear Physics (Munich), Vol. 1, 517. Amsterdam: North-Holland/American Elsevier.
[4]. Szytula, A., & Leciejewicz, J. (1989). Handbook on the Physics and Chemistry of Rare Earths, Vol. 12 (K. A. Gschneidner Jr & L. Erwin, Eds.), 133. Amsterdam: Elsevier.
[5]. Kuhn, T. (1998). Density matrix theory of coherent ultrafast dynamics. In Theory of Transport Properties of Semiconductor Nanostructures (Electronic Materials Vol. 4, E. Schöll, Ed.), 173–214. London: Chapman and Hall.
[6]. Magdacy Jerjes, A., Zeki Ablahd, N., Yousif Dawod, A., & Fakhrulddin Abdulqader, M. (2023). Detect malicious web pages using naive Bayesian algorithm to detect cyber threats. Wireless Personal Communications, 1-13.
[7]. Zhang, H., Jiang, L., & Webb, G. I. (2023). Rigorous non-disjoint discretization for naive Bayes. Pattern Recognition, 140, 109554.
[8]. Talaei Khoei, T., & Kaabouch, N. (2023). A comparative analysis of supervised and unsupervised models for detecting attacks on intrusion detection systems. Information, 14(2), 103.
[9]. Dhiman, R. (2023). Electroencephalogram channel selection based on Pearson correlation coefficient for motor imagery-brain-computer interface. Measurement: Sensors, 25, 100616.
[10]. Chicco, D., & Jurman, G. (2023). The Matthews correlation coefficient (MCC) should replace the ROC AUC as the standard metric for assessing binary classification. BioData Mining, 16(1), 4.
Cite this article
Luo,L.;Liu,T. (2024). Integrating advanced principal component analysis into naive bayes for enhanced classification performance. Advances in Operation Research and Production Management,3,27-31.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Journal:Advances in Operation Research and Production Management
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Mushtaq, Z., et al. (2023). Effective kernel‐principal component analysis based approach for Wisconsin breast cancer diagnosis. Electronics Letters, 59(2), e212706.
[2]. Yesilkaya, B., et al. (2023). Principal component analysis and manifold learning techniques for the design of brain-computer interfaces based on steady-state visually evoked potentials. Journal of Computational Science, 68, 102000.
[3]. Caplar, R., & Kulisic, P. (1973). Proc. Int. Conf. on Nuclear Physics (Munich), Vol. 1, 517. Amsterdam: North-Holland/American Elsevier.
[4]. Szytula, A., & Leciejewicz, J. (1989). Handbook on the Physics and Chemistry of Rare Earths, Vol. 12 (K. A. Gschneidner Jr & L. Erwin, Eds.), 133. Amsterdam: Elsevier.
[5]. Kuhn, T. (1998). Density matrix theory of coherent ultrafast dynamics. In Theory of Transport Properties of Semiconductor Nanostructures (Electronic Materials Vol. 4, E. Schöll, Ed.), 173–214. London: Chapman and Hall.
[6]. Magdacy Jerjes, A., Zeki Ablahd, N., Yousif Dawod, A., & Fakhrulddin Abdulqader, M. (2023). Detect malicious web pages using naive Bayesian algorithm to detect cyber threats. Wireless Personal Communications, 1-13.
[7]. Zhang, H., Jiang, L., & Webb, G. I. (2023). Rigorous non-disjoint discretization for naive Bayes. Pattern Recognition, 140, 109554.
[8]. Talaei Khoei, T., & Kaabouch, N. (2023). A comparative analysis of supervised and unsupervised models for detecting attacks on intrusion detection systems. Information, 14(2), 103.
[9]. Dhiman, R. (2023). Electroencephalogram channel selection based on Pearson correlation coefficient for motor imagery-brain-computer interface. Measurement: Sensors, 25, 100616.
[10]. Chicco, D., & Jurman, G. (2023). The Matthews correlation coefficient (MCC) should replace the ROC AUC as the standard metric for assessing binary classification. BioData Mining, 16(1), 4.









