1. Introduction
Diabetes is a serious chronic disease whose prevalence is increasing at an alarming rate. To prevent this disease, researchers are actively developing predictive models for detecting and analyzing the condition. Studies have been conducted to develop diabetes prediction models specifically for women based on some of their characteristics including blood glucose levels, skin thickness, number of pregnancies, etc. This in turn predicts whether they are likely to develop diabetes or not. Early detection and prediction of diabetes in women is significant because of the increased risk of diabetes-related complications during pregnancy.
Nowadays, several studies have explored predictive models for diabetes. Jayanthi et al. reviewed various predictive models used in healthcare, specifically mentioning models for diabetes prediction [1]. They talked about how data mining and machine learning may be used to create precise prediction models. Machine learning is a group of algorithms that, without outside programming, allow software programs to forecast more accurate results. Applying computational methods that utilize statistical analysis to predict outcomes from input data and update the output data is the fundamental tenet of machine learning (ML) [2]. In the field of machine learning structure, Random Forest (RF) is one of the most effective and popular models [3]. For many supervised approaches, notably decision trees, it can enhance predictions. A group of classifiers with low bias and large variance are the end result. The ensemble approach then lowers the variance by averaging the resulting trees to calculate predictions [4]. Southern et al. studied the validity of using administrative databases to identify diabetes patients and discussed how linking laboratory data can improve accuracy [5]. Based on the development of new algorithms, the collection of large data, and the expansion of computer processing power. Artificial Intelligence techniques such as neuron networks have been booming globally [6]. Researcher online messages made on social media have recently been used to gather biological characteristics of diabetes patients, applying closed loops, insulin pumps, or continuous blood glucose monitors, for instance. A better and more thorough characterization of the many kinds and presentations of diabetes may be possible with the use of data and techniques like unsupervised learning [7]. The French National Health Insurance Information System (SNDS) uses three diabetes case definition algorithms to identify diabetics. All of those models functioned effectively, in the identification research conducted by S. Fuentes, et al [8]. Based on the Pima Indian diabetes dataset, Lakhwani et al. employed an artificial neural network to create a diabetes predicting model [9]. The current study will build upon this previous work by developing a model that specifically targets predicting the risk of females developing diabetes. This targeted model has the potential to enable early intervention and management strategies that could help reduce the burden of diabetes and its complications in women.
The goal of this research is to utilize ML to build possible forecasting algorithms to effectively assess the likelihood of diabetes in women. Specifically, first, data preprocessing is used to address missing values and outliers in the dataset. We replace the zero values of certain features such as "blood glucose" and "blood pressure" with the median values of the corresponding features. This step reduces the impact of less reliable data on model performance. Second, we introduce techniques for building predictive models, including Gaussian Naive Bayes (GNB), Support Vector Machine (SVM), and RF. They have superior robustness compared to other traditional algorithms. Third, we perform model screening. The optimal data segmentation strategy is determined by comparing the prediction performance of the models under different training-testing segmentations. Experimental findings indicate that the RF algorithm outperforms the other two models in prediction accuracy on the dataset. The introduction of machine learning to construct predictive models in this study enables physicians to predict the risk of diabetes in patients earlier. Additionally, this facilitates timely intervention in the development process to effectively prevent and control diabetes.
2. Methodology
2.1. Dataset description and preprocessing
This project's basis is the Diabetes Database, which was produced by the National Institute of Diabetes and Digestive and Kidney Diseases and is now available to the general public on Kaggle [10]. Eight medical predictor factors, including the number of pregnancies, BMI, insulin level, and age, are included in the dataset. A binary outcome variable is also included, indicating the presence or absence of diabetes.
During preprocessing, integrity, and reliability are prioritized due to their direct impact on the model's predictive performance. Zero values are assigned to characteristics including blood pressure, thickness of the skin, glucose, and BMI, which are likely placeholders for missing data or inaccuracies. These are substituted with the median value of their respective columns to avoid data skewness. Following this cleaning step, the dataset is then partitioned into training, validation, and testing sets to serve different phases: model training, hyperparameters tuning, model selection, and final evaluation.
2.2. Proposed approach
The main aim of this research revolves around the creation of a user-friendly, easily accessible tool that employs machine learning algorithms to enable individuals to evaluate their risk of developing diabetes. The tool is built on three distinct machine learning algorithms: GNB, SVM, and RF. The functionality of this tool will allow it to deliver an initial risk assessment to individuals, thereby potentially encouraging those with higher risk profiles to seek professional medical advice earlier and potentially deter the onset of severe health complications associated with diabetes. The processing is shown in the Figure 1.
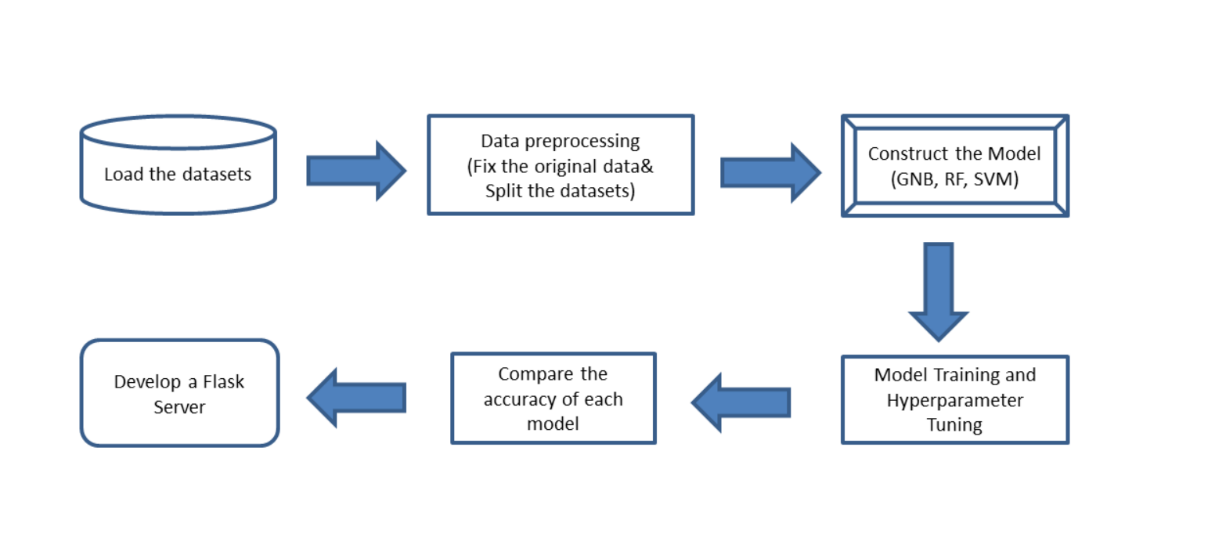
Figure 1. The methodology progress on machine learning of diabetes prediction.
2.2.1. GNB.GNB is a probabilistic classifier rooted in the Naive Bayes family, particularly designed for continuous data types. The foundational principle behind GNB is its assumption that each feature, when segmented under a specific class, will follow a Gaussian distribution. The algorithm operates on a straightforward framework, as follows,
\( P(X=x|C=c)=\frac{1}{\sqrt[]{2πσ}}{e^{\frac{-{(x-μ)^{2}}}{2{σ^{2}}}}} \) (1)
Firstly, it computes the likelihood of a certain feature X under the umbrella of a given class C using the Gaussian probability density function. This calculation depends on parameters such as the mean (μ) and standard deviation (σ) for that feature within that class. Secondly, leveraging Bayes' theorem, GNB calculates the posterior probabilities for every class [11]. During the classification phase, the algorithm examines these posterior probabilities and assigns the class with the highest value to the data instance.
2.2.2. SVM. SVM is recognized as a potent supervised learning algorithm primarily tailored for the domain of classification. The core principle driving SVM is the search for an optimal hyperplane, one that adeptly divides different classes within a dataset. The emphasis on optimizing the gap, commonly referred to as the spacing between the hyperplane and the adjacent data points from both categories, is a pivotal aspect of the system [2]. Addressing non-linearly separable datasets, SVM introduces kernel functions such as the linear or radial basis function. These kernels transform the data's feature space, equipping the algorithm to navigate complex data patterns and effectuate precise classifications.
2.2.3. RF. The RF technique is an ensemble learning method, gaining its strength from aggregating the outcomes of numerous decision trees. Instead of anchoring its decisions on a singular tree, RF capitalizes on the collective insights of multiple trees, thus nullifying individual biases. Its framework involves the cultivation of multiple trees, each trained on bootstrapped subsets of data, and further utilizing a randomized set of features [3]. This strategy infuses variability among the trees, fortifying the model's resilience. When classifying or predicting, each tree within the forest renders its verdict. In classification scenarios, it manifests as a vote for a specific class, and in regression, it offers a continuous prediction. RF consolidates these outputs: for classifications, the majority vote becomes the final decision, while for regression tasks, it averages the tree predictions. This collective methodology empowers RF to offer consistent and dependable results across a plethora of scenarios.
2.2.4. Model training and hyperparameter tuning. Each machine learning algorithm incorporated in this project was chosen for its unique strengths. The GNB algorithm is known for its efficiency and simplicity, making it particularly suitable for larger datasets. In contrast, the SVM excels in managing high-dimensional data and modeling intricate, nonlinear relationships. RF, an ensemble learning method, is inherently flexible and robust against overfitting, even when dealing with high-dimensional data. The integration of these diverse algorithms creates a versatile and powerful predictive system.
Following the construction of the models, the next critical phase was their training. This process was contingent on the algorithm in use; for instance, GNB, due to its simplicity, required relatively less tuning. Conversely, SVM performance is highly sensitive to the kernel parameter, thereby benefiting from careful tuning to effectively model non-linear data. The RF model requires extensive tuning of hyperparameters such as the number of decision trees and the maximum depth of these trees to prevent overfitting and ensure reliable performance. The dataset was further divided into training (60%), validation (20%), and testing (20%) subsets to maintain a robust equilibrium between training the models and validating their performance with unseen data.
2.2.5. Flask server development. The final stage in the proposed approach involves leveraging Flask, a Python-based microweb framework, to convert the trained machine learning models into an accessible and user-friendly web application. Flask's minimalistic and flexible nature allows for rapid prototyping and deployment of web-based services. For this project, it acts as a bridge between the sophisticated machine learning algorithms and users, enabling them to assess their risk of diabetes using a simple interface. The Flask server was designed to take user inputs (medical parameters) through a web interface, pass them to the trained models, and return the prediction results. This step provides users immediate access to diabetes risk predictions based on their health parameters, emphasizing the potential impact of this research on preventive healthcare.
3. Result and discussion
Model training, optimization, and performance evaluation revealed important insights into the machine learning models' ability to effectively classify the given dataset. The process of model optimization comprised several distinct stages.
The initial focus is on optimizing the RF model through hyperparameter tuning. This involved varying parameters such as the number of estimators, the maximum depth, and the minimum sample leaves. As shown in Figure 2 and Figure 3, the tuning of each of these parameters contributed to the model's performance, with optimal levels found for each. The Receiver Operating Characteristic - Area Under the Curve (ROC-AUC) score is at its highest when the number of estimators is 60, the maximum depth is 5, and the minimum sample leaf is 1, indicating an optimized model configuration.
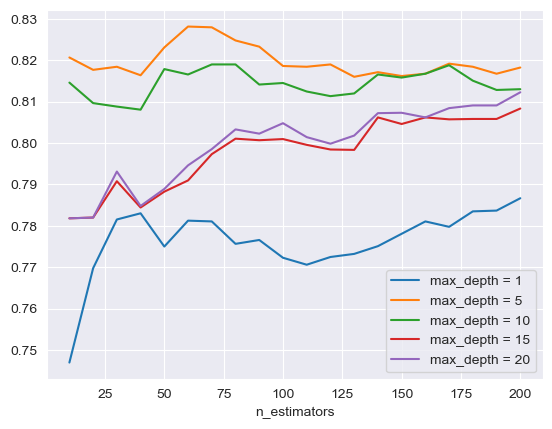
Figure 2. The performance comparison of different depths. max_depth represents the maximum depth of RF hyperparameter tuning.
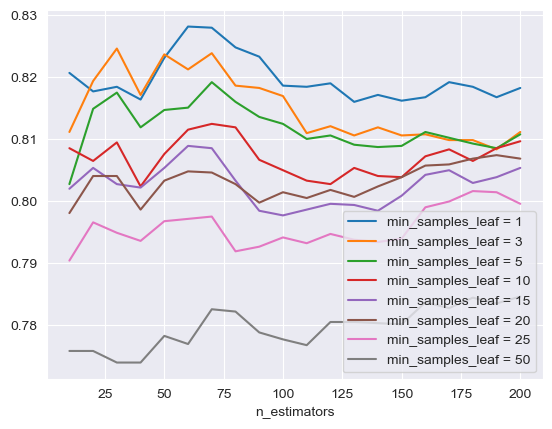
Figure 3. The performance comparison of different leaves. min_sample_leaf represents the minimum sample leaf in RF hyperparameter tuning.
Once optimized, along with other trained models, the Random Forest model's performance is assessed. This is done using the ROC-AUC score, a reliable metric for model performance in classification problems. As seen in Table.1, while all models demonstrated appreciable performance, certain models achieved a higher ROC-AUC score, suggesting superior performance in accurately classifying the instances of the dataset.
Table 1. Chart of different data accuracy.
Accuracy(probability) | NB | SVM | RF |
Training Data (before tuning) | 0.7639 | 0.7949 | 0.7818 |
Training Data | 0.7639 | 0.8290 | 0.8281 |
Test Data | 0.8483 | 0.8599 | 0.8724 |
In a practical application context, the best performing model is deployed in a Flask application, shown in Figure 4. This interactive interface allows users to input data and receive model predictions, thus extending the model's use beyond an analytical context to a practical, user-centric application. Future work may involve improving this application for enhanced user experience, along with integrating additional models for comparative prediction. Through the above analysis and experiments, the expressiveness of various machine learning models on the classification problem of a given data set is clarified. These experiments show that with proper parameter tuning, random forest models can achieve optimal expressiveness. Meanwhile, the performance comparison of different models reveals that some models have higher accuracy in the ROC-AUC score. Furthermore, the model is scaled for practical use by deploying the best-performing model in a Flask application.
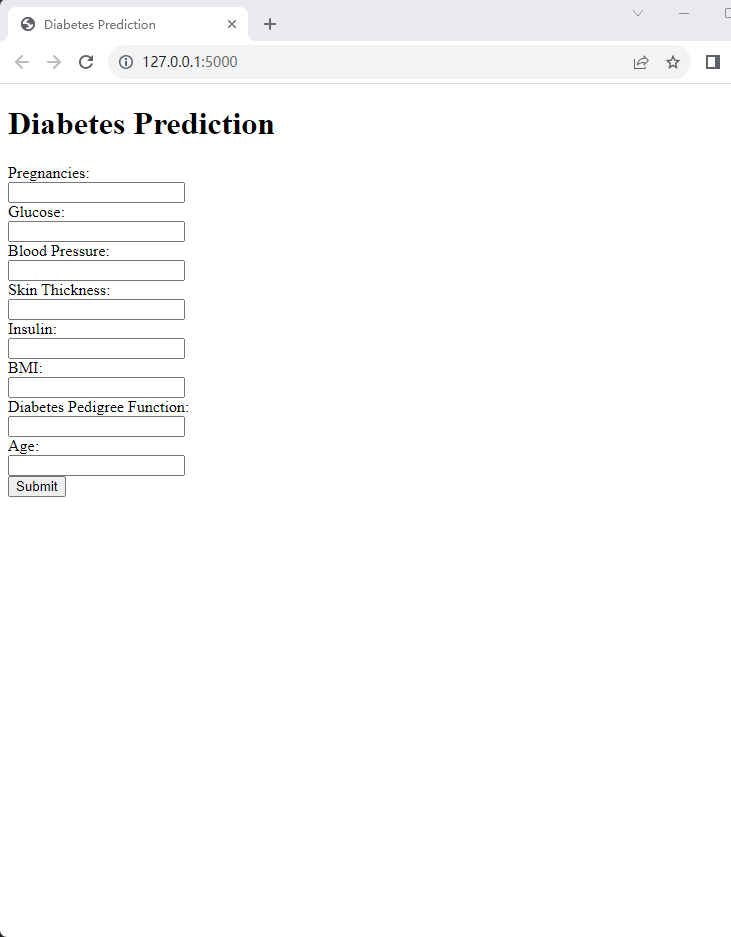
Figure 4. Flask application web page.
4. Conclusion
In this study, we introduce algorithms such as GNB, SVM, and RF to construct predictive models to assess the likelihood of diabetes in women. Missing values and outliers in the dataset were excluded in the preprocessing stage. The performance of different models is compared and analyzed based on ROC-AUC scores while parameter tuning. Additionally, to extend the practical application, the best model is incorporated into the Flask application for practical use. The experimental results demonstrate the superior robustness of the tuned random forest model compared to other traditional algorithms. Specifically, with the proposed approach, the model not only improves the accuracy but also highlights the critical role of specific features in influencing classification. In the future, the main goal of the next phase of research is to delve into feature engineering to reveal the potential relationships between features and their combined impact on prediction accuracy.
Authors contribution
All the authors contributed equally and their names were listed in alphabetical order.
References
[1]. Jayanthi N Vijaya B B Sambasiva R N 2017 Survey on clinical prediction models for diabetes prediction Journal of Big Data 4(1): pp 1-15
[2]. Jaber Q Saeid B Kourosh E Yasaman A 2022 A survey of machine learning in kidney disease diagnosis Machine learning with Applications 10: p 100418
[3]. Breiman L 2001 Random forests Machine Learning 45(1): 5-32
[4]. Hastie T Tibshirani R Friedman J 2009 The Elements of Statistical Learning: Data Mining, Inference, and Prediction Springer Science & Business Media
[5]. Southern D A Roberts B Edwards A Dean S Norton P Svenson L W Ghali W A 2010 Validity of administrative data claim-based methods for identifying individuals with diabetes at a population level Canadian Journal of Public Health pp 61-64
[6]. Yang X Wang M Zhou Y 2021 The development trend of artificial intelligence in medical: A patentometric analysis Artificial Intelligence in the Life Sciences 1: p 100006
[7]. Guy F 2021 Challenges and perspectives for the future of diabetes epidemiology in the era of digital health and artificial intelligence Diabetes Epidemiology and Management 1: p100004
[8]. Sonsoles F Emmanuel C Laurence M Anne F Pascale B Marcel G Sandrine F and CONSTANCES-Diab Group 2019 Identifying diabetes cases in health administrative databases: a validation study based on a large French cohort International Journal of Public Health 64(3): pp 441-450
[9]. Lakhwani G M Patel S C Rajendra S Prajapati L B 2020 Prediction of diabetes using neural network Materials Today: Proceedings 21: pp 1470-1474
[10]. Khare A D 2022 Diabetes dataset https://www.kaggle.com/datasets/akshaydattatraykhare/diabetes-dataset
[11]. Kamel D Abdulah J M 2019 Cancer Classification Using Gaussian Naive Bayes Algorithm 2019 International Engineering Conference (IEC) Erbil pp 165-170
Cite this article
Han,Y.;Li,Q.;Lou,J.;Zhang,J. (2024). Prediction of diabetes progress based on machine learning approach. Applied and Computational Engineering,37,45-51.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 2023 International Conference on Machine Learning and Automation
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Jayanthi N Vijaya B B Sambasiva R N 2017 Survey on clinical prediction models for diabetes prediction Journal of Big Data 4(1): pp 1-15
[2]. Jaber Q Saeid B Kourosh E Yasaman A 2022 A survey of machine learning in kidney disease diagnosis Machine learning with Applications 10: p 100418
[3]. Breiman L 2001 Random forests Machine Learning 45(1): 5-32
[4]. Hastie T Tibshirani R Friedman J 2009 The Elements of Statistical Learning: Data Mining, Inference, and Prediction Springer Science & Business Media
[5]. Southern D A Roberts B Edwards A Dean S Norton P Svenson L W Ghali W A 2010 Validity of administrative data claim-based methods for identifying individuals with diabetes at a population level Canadian Journal of Public Health pp 61-64
[6]. Yang X Wang M Zhou Y 2021 The development trend of artificial intelligence in medical: A patentometric analysis Artificial Intelligence in the Life Sciences 1: p 100006
[7]. Guy F 2021 Challenges and perspectives for the future of diabetes epidemiology in the era of digital health and artificial intelligence Diabetes Epidemiology and Management 1: p100004
[8]. Sonsoles F Emmanuel C Laurence M Anne F Pascale B Marcel G Sandrine F and CONSTANCES-Diab Group 2019 Identifying diabetes cases in health administrative databases: a validation study based on a large French cohort International Journal of Public Health 64(3): pp 441-450
[9]. Lakhwani G M Patel S C Rajendra S Prajapati L B 2020 Prediction of diabetes using neural network Materials Today: Proceedings 21: pp 1470-1474
[10]. Khare A D 2022 Diabetes dataset https://www.kaggle.com/datasets/akshaydattatraykhare/diabetes-dataset
[11]. Kamel D Abdulah J M 2019 Cancer Classification Using Gaussian Naive Bayes Algorithm 2019 International Engineering Conference (IEC) Erbil pp 165-170









