1. Introduction
As medical technology, especially medical imaging technology, rapidly advances worldwide, early diagnosis and treatment of brain diseases have become critical topics in modern medical research. Brain diseases, particularly tumors, pose serious threats to patient health due to their complexity. Accurate diagnosis and timely treatment are highly demanded. Correct identification and classification of brain tumors are crucial for treatment option selection and prognostic evaluation. However, the high heterogeneity of brain diseases makes traditional diagnostic methods slow and limited. Hence, an efficient, accurate automated diagnostic system is necessary.
Deep learning, especially Convolutional Neural Networks (CNN), demonstrates potential in medical image analysis, offering new methods for automatic detection and classification of brain tumors. By learning from a vast amount of medical images, deep learning models can automatically extract and recognize features within the images, aiding doctors in making more accurate tumor diagnosis and treatment decisions. Recent years have seen various deep learning architectures[1-3], like AlexNet[4], VGG[5], and ResNet[6], being proposed and applied in medical image analysis, significantly driving the automation of medical diagnosis.
This study aims to explore and optimize the application of deep learning models in the classification of brain CT images, specifically for the recognition of aneurysms, cancer, and malignant tumors. We employed an improved ResNet-18 model, chosen both for its exceptional feature extraction capabilities and for its relatively lower computational demand, suitable for processing high-resolution medical images. By refining the model's structure and optimization, we expect to maintain high accuracy while improving the model's adaptability and generalization capability for small datasets.
2. Relevant Work
In recent years, the application of deep learning models in medical CT image classification has garnered attention. Before this, researchers had explored and applied various deep learning models for image classification tasks. Besides ResNet-18, other models have also been employed for similar tasks.
Alex Krizhevsky, Ilya Sutskever, and Geoffrey Hinton introduced the AlexNet model in 2012 at NIPS.[4] As a milestone in deep learning, AlexNet features effective feature extraction capabilities with wide application prospects. However, it suffers from significant computing and memory demands, and a tendency for overfitting.
Karen Simonyan and Andrew Zisserman presented the VGG model in 2014 on arXiv. [5]The VGG model, with its simple yet effective architecture, modular design, and lower parameter count, has advantages. Yet, it also deals with high computing and memory demands, a tendency for overfitting, lengthy training times, and unsuitability for real-time applications.[7]
Kaiming He, Xiangyu Zhang, Shaoqing Ren, and Jian Sun introduced the ResNet model at the IEEE Conference on Computer Vision and Pattern Recognition (CVPR) in 2015. [6]ResNet addresses the issues of gradient vanishing and network degradation in traditional deep neural networks through residual learning, allowing deeper networks to be trained more effectively and efficiently.
The dataset for recognizing types of brain diseases comprises three main diseases: aneurysms, cancer, and malignant tumors, including 259 images subdivided into 84 aneurysm, 91 cancer, and 84 malignant tumor images, aiming to classify and diagnose effectively through deep learning models. For this purpose, the images are split into subsets: 208 for model training, 28 for validation, and 27 for final testing. Each image maintains a resolution of 512×512 pixels, having 262,144 pixels and associated with three different category labels for classification tasks. Figure 1. displays 20 random images from the test set.
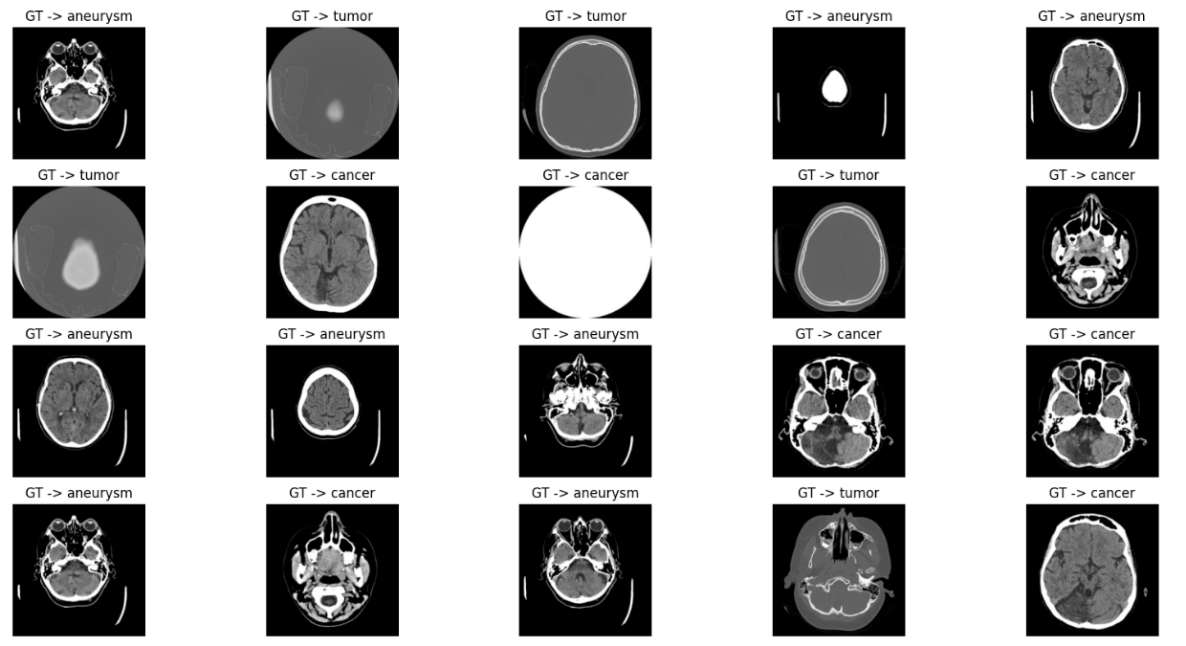
Figure 1. 20 random images
3. Methodology
Convolutional Neural Networks (CNN), a unique neural network architecture, automatically learn hierarchical features of data through their distinctive layer structure. CNN's operation includes using convolutional layers to extract spatial features, introducing non-linearity with activation functions, reducing computational load and preventing overfitting with pooling layers, and finally performing classification or regression through fully connected layers. These characteristics make CNNs especially suitable for vision-related tasks such as image and video recognition, image classification, and object detection, now widely applied in autonomous driving, facial recognition, and medical image analysis domains. ResNet-18, introduced by Kaiming He and others in 2015 and demonstrated remarkable performance in the ImageNet large-scale visual recognition challenge, is a smaller version of the deep convolutional neural network. By adopting the concept of residual learning, ResNet efficiently addresses the problems of gradient vanishing and expression bottlenecks encountered during the training of deep neural networks. Residual blocks pass information directly across layers through shortcut connections, enabling the network to learn identity mapping more easily.[10]
Compared to traditional neural network models (Figure 2.), ResNet opts not to force a few layers of hierarchical structure to learn the desired underlying mapping directly but to have these layers learn residual mappings, as shown in formula (1). This means if the optimal solution requires the identity mapping, driving the residuals toward zero is more straightforward and easier than fitting the identity mapping through a series of non-linear layers. This architecture ensures increased depth does not degrade learning capability and performance.
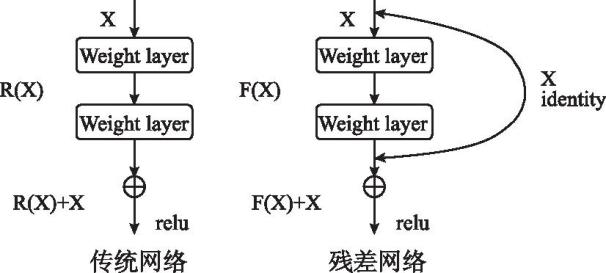
Figure 2. traditional neural network models
\( H(x)=F(x)+x\ \ \ (1) \)
ResNet50 is a classic deep learning model belonging to the CNN category. It extracts image features by stacking different levels of convolution and residual blocks. The structure of ResNet-18 is relatively simple, consisting of multiple residual blocks and pooling layers. It comprises 18 convolutional layers, including 16 convolutional layers and 2 fully connected layers. The overall framework of this paper's model is as depicted in table1.
Table 1. overall framework of this paper's model
Layer | Output Shape | Param | Description |
Input channels | (None, 224, 224, 3) | images with RGB channels | |
Conv1(Conv2D) | (None, 112, 112, 64) | 9472 | Convolutiona layer, 64 filters, 7x7 kernel, stride 2 |
BN1 | (None, 112, 112, 64) | 256 | Batch normalization |
ReLU | (None, 112, 112, 64) | ReLU activation | |
MaxPooling2D window, stride2 | (None, 56, 56, 64) | Max pooling with3x3 | |
ResBlock1_1 | (None, 56, 56, 64) | First residual block | |
ResBlock1_2 | (None, 56, 56, 64) | Second residual block | |
ResBlock2_1 | (None, 28, 28, 128) | Third residual block | |
ResBlock2_2 | (None, 28, 28, 128) | Fourth residual block | |
ResBlock3_1 | (None, 14, 14, 256) | Fifth residual block | |
ResBlock3_2 | (None, 14, 14, 256) | Sixth residual block | |
ResBlock4_1 | (None, 7, 7, 512) | Seventh residual block | |
ResBlock4_2 | (None, 7, 7, 512) | Eighth residual block | |
GlobalAvgPooling2D | (None, 512) | Global average pooling | |
Dense units | (None, 3) | 513000 | Fully connected layer with 3 |
Output | (None, 3) | Output layer |
To address the high resolution of brain CT images and the characteristics of small datasets, this paper optimizes the model with the following strategies:
Adjustment of Convolutional Layer Parameters: Normalizing images allows the model to more effectively process data from different sources, thus enhancing training efficiency and model stability.
Processing Techniques: Normalization helps the model better handle images from various sources, improving the efficiency and stability of model training.
Model Training Strategy: Given the common challenge of small-scale medical image datasets and high model complexity, we enhance the model's generalization ability and overall performance by selecting appropriate loss functions and optimization strategies.
4. Model Training
Begin by loading the pre-trained ResNet-18 model, then fine-tune it for the new classification task. Training commences with initialized loss and accuracy metrics, obtaining predictions through forward propagation, followed by loss computation (cross-entropy loss function), reflecting the model's current performance status. The training process monitors the number of correct predictions and cumulative loss in each batch, using these metrics to evaluate and enhance model performance. Model parameters are continuously optimized through backpropagation and the Adam optimizer, allowing the model to better fit the training data.
At the end of each training epoch, the model undergoes validation on an independent set to assess its classification ability on unknown data.
If the accuracy on the validation set surpasses the previously saved best accuracy, we update the best accuracy and save the current model weights.
5. Result Analysis
Table 2. briefly summarizes the model's training and validation performance across the first 10 epochs. Results show the model's training performance improving with increasing epochs, with training loss gradually reducing and the accuracy climbing to a final rate of 0.956. However, model performance on the validation set fluctuated to some extent. The validation accuracy was higher in the initial epochs but saw a slight dip thereafter, eventually stabilizing at 0.960. This indicates potential overfitting, especially in later training phases, potentially linked to the smaller dataset size.
Table 2. summarizes the model's training and validation performance
Epoch | Train Loss | Train Accuracy | Validation Loss | Validation Accuracy |
1 | 0.830 | 0.696 | 0.511 | 0.920 |
2 | 0.181 | 0.957 | 0.196 | 0.920 |
3 | 0.045 | 0.986 | 0.278 | 0.880 |
4 | 0.152 | 0.957 | 0.088 | 0.960 |
5 | 0.034 | 0.990 | 0.105 | 0.960 |
6 | 0.025 | 0.981 | 0.116 | 0.960 |
7 | 0.024 | 0.986 | 0.108 | 0.960 |
8 | 0.021 | 0.986 | 0.148 | 0.960 |
9 | 0.034 | 0.981 | 0.089 | 0.960 |
10 | 0.021 | 0.986 | 0.123 | 0.960 |
6. Prediction Results
Using the trained model to infer on the given test dataset, comparing inference results with actual labels. A certain number of image samples were randomly selected for display, each title including the true category and the model's predicted category, as shown in figure 3.
Results indicate that for accurately predicted images, titles are displayed in green; for incorrectly predicted ones, in red. This visual method allows an intuitive understanding of the model's performance on the test dataset, identifying potential classification errors and providing a direct assessment of model performance.
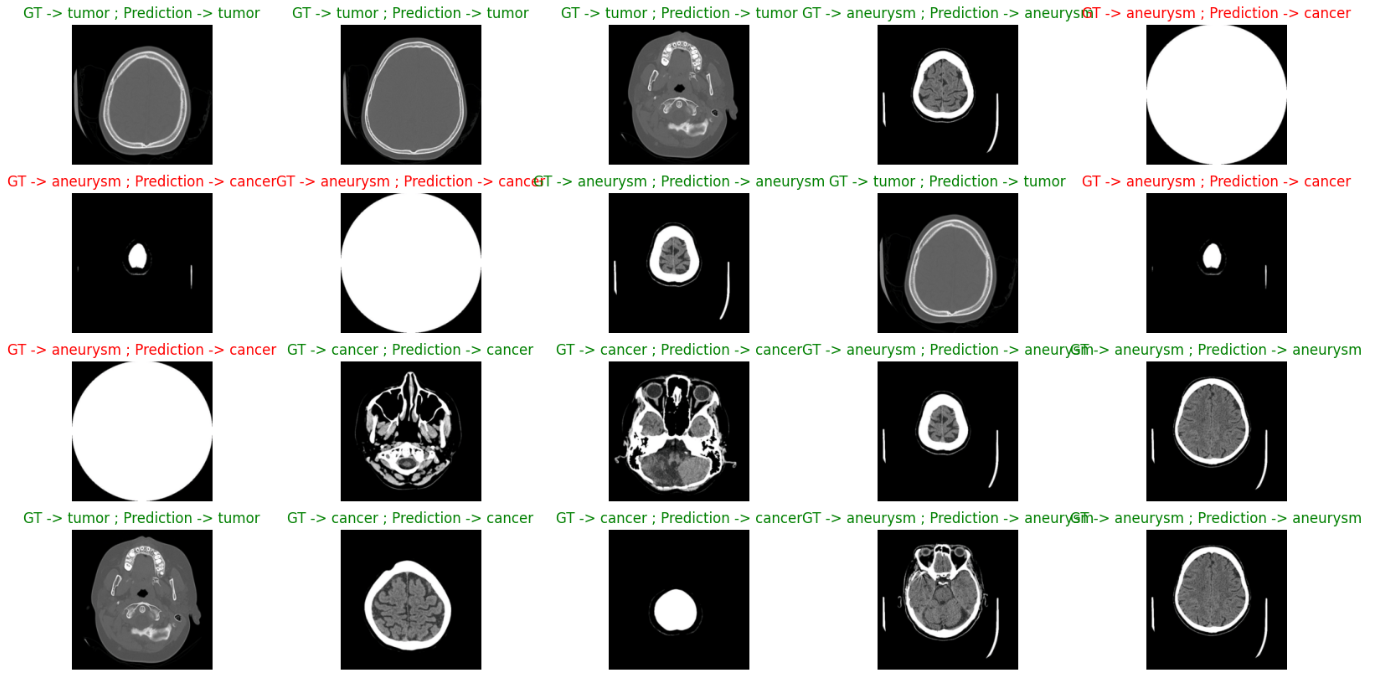
Figure 3. true category and predicted category
7. Summary
This paper utilizes an optimized ResNet-18 model for classifying aneurysms, cancer, and malignant tumors in brain CT images. To adapt to the high-resolution characteristics of medical images, the model's feature extraction layer was carefully designed. Throughout the training process, the model's generalization ability for small datasets was enhanced by employing a pre-trained ResNet-18 model, fine-tuning for the brain disease image classification need, and using an Adam optimizer and cross-entropy loss function, updating model parameters through back-propagation.
The use of deep learning technology, especially the improved ResNet-18 model, has achieved fairly accurate predictions in the automatic classification task of brain CT images. Despite potential issues of overfitting and accuracy fluctuation, these results still demonstrate the strong potential of deep learning in automatic medical image analysis. Future work may focus on the following areas:
Development of Interpretability Tools: Developing and integrating new visualization tools and interpretive algorithms to allow medical professionals to intuitively understand the model's decision-making process, increasing the credibility of medical diagnoses.
Clinical Validation: Further validating the model's accuracy and practicality by comparing with actual clinical diagnostic results, focusing on the consistency between model predictions and doctors' diagnoses to ensure the model's practical value.
Adaptation to Different Medical Environments: Considering the differences in CT scanning equipment and image acquisition in different medical environments, future work will explore how to adjust the model to accommodate these changes, ensuring high performance in various real-world scenarios.
References
[1]. Ruuskanen, O., Lahti, E., Jennings, L. C., & Murdoch, D. R. (2011). Viral pneumonia. The Lancet, 377(9773), 1264-1275. https://doi.org/10.1016/S0140-6736(10)61459-6
[2]. Xie, J., & Zhang, K. (2024). SOSNet: A non-small cell lung cancer CT image segmentation model with asymmetric encoder-decoder structure. *Journal of Electronics*, 1-15. https://kns-cnki-net.webvpn.ybu.edu.cn/kcms/detail/11.2087.TN.20240311.1736.016.html.
[3]. Yang, W. (2024). Research on liver tumor segmentation method of CT images based on deep learning [Doctoral dissertation, Hunan University of Science and Technology]. DOI: 10.27738/d.cnki.ghnkd.2022.000728.
[4]. Krizhevsky, A., Sutskever, I., & Hinton, G. E. (2012). ImageNet classification with deep convolutional neural networks. In Advances in Neural Information Processing Systems (pp. 1097-1105).
[5]. Simonyan, K., & Zisserman, A. (2014). Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556.
[6]. He, K., Zhang, X., Ren, S., & Sun, J. (2016). Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 770-778).
[7]. Marriam, N., Sumera, S., Momina, M., et al. (2024). COVID-ECG-RSNet: COVID-19 classification from ECG images using swish-based improved ResNet model. Biomedical Signal Processing and Control, 89105801-.
[8]. Li, J., Liu, H., Li, Y. et al. (2024). Image recognition based on THGS algorithm optimized ResNet-18 model. Journal of Jilin University (Engineering and Technology Edition), 1-10. https://doi.org/10.13229/j.cnki.jdxbgxb.20230775.
Cite this article
Zhang,G. (2024). A ResNet based transfer learning pulmonary medical image disease detection system. Applied and Computational Engineering,88,190-195.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 6th International Conference on Computing and Data Science
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Ruuskanen, O., Lahti, E., Jennings, L. C., & Murdoch, D. R. (2011). Viral pneumonia. The Lancet, 377(9773), 1264-1275. https://doi.org/10.1016/S0140-6736(10)61459-6
[2]. Xie, J., & Zhang, K. (2024). SOSNet: A non-small cell lung cancer CT image segmentation model with asymmetric encoder-decoder structure. *Journal of Electronics*, 1-15. https://kns-cnki-net.webvpn.ybu.edu.cn/kcms/detail/11.2087.TN.20240311.1736.016.html.
[3]. Yang, W. (2024). Research on liver tumor segmentation method of CT images based on deep learning [Doctoral dissertation, Hunan University of Science and Technology]. DOI: 10.27738/d.cnki.ghnkd.2022.000728.
[4]. Krizhevsky, A., Sutskever, I., & Hinton, G. E. (2012). ImageNet classification with deep convolutional neural networks. In Advances in Neural Information Processing Systems (pp. 1097-1105).
[5]. Simonyan, K., & Zisserman, A. (2014). Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556.
[6]. He, K., Zhang, X., Ren, S., & Sun, J. (2016). Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition (pp. 770-778).
[7]. Marriam, N., Sumera, S., Momina, M., et al. (2024). COVID-ECG-RSNet: COVID-19 classification from ECG images using swish-based improved ResNet model. Biomedical Signal Processing and Control, 89105801-.
[8]. Li, J., Liu, H., Li, Y. et al. (2024). Image recognition based on THGS algorithm optimized ResNet-18 model. Journal of Jilin University (Engineering and Technology Edition), 1-10. https://doi.org/10.13229/j.cnki.jdxbgxb.20230775.









