1. Introduction
Brain cancer is a cancer that can be directly identified without specific symptoms, but brain imaging is a useful aid to help doctors diagnose and distinguish brain cancer conditions [1]. But a doctor's diagnosis often requires years of clinical experience. Missed diagnosis and misdiagnosis can lead to patients not receiving timely treatment and increase the death rate of brain cancer. The artificial intelligence (AI) model can effectively help medical examiners quickly screen possible patients, thereby improving medical efficiency and effectively saving time for specialists. The accuracy of the AI model itself is no less than that of a doctor's diagnosis, and it can effectively subdivide diseases for precise treatment.
According to the summary of La ´s Silva Santana et al., most researchers used support vector machine (SVM) and random forest for model construction. Most sensitive and specificity are above 90% [2]. This shows that the accuracy of the current mainstream models is not very different. Some researchers try to improve the training speed of the model by effectively preprocessing the data. B. Shyamala has effectively improved the accuracy of the model by adding an anisotropic diffusion filter to the picture [3]. Sarah Ali Abdelaziz Ismael used residual networks to achieve more than 99% accuracy on the open data set [4]. By improving the Hunger Games Search Algorithm, Marwa M. Emam also obtained high accuracy and proved its application potential in the field of brain cancer classification [5]. P.S. Tejashwini, on the other hand, uses a multi-classification network to achieve a more accurate and subdivided diagnosis of brain cancer. The classification efficiency of the model was improved by subdividing the brain tumor tissue and normal tissue [6]. SK Rajeev first separated the skull features, and then tried to extract the characteristics of the tumor region to distinguish it more effectively [7].
To improve the ease of model learning while maintaining the ability to efficiently distinguish between different types of brain cancer, this study leverages data from the Kaggle database and employs a two-pronged approach. Firstly, the study evaluates the performance of basic convolutional neural network (CNN) models to determine their capability in achieving a satisfactory level of classification. The analysis focuses on the impact of various convolutional layers and kernel sizes on receptive field extraction. After experimentation, a model configuration with four convolutional layers and three kernels per layer was found to be optimal, allowing the model to achieve over 90% accuracy even with a limited dataset. Secondly, to enhance learning efficiency and reduce the computational resources required, the study explores model transfer techniques, specifically employing EfficientNet2 for transfer learning. This approach is particularly advantageous during the early stages of training, as the deep convolution processes in EfficientNet2 progress more slowly, thereby minimizing resource consumption [8]. The study also uses smaller-sized images for training, successfully maintaining accuracy without significant precision loss.
A crucial aspect of this research is optimizing the combination of Mobile Convolution (MBConv) and Fusion-MBConv within the model. By leveraging neural networks, the study quickly identifies the best configuration, reducing the time needed for parameter tuning. This optimization not only accelerates the recognition speed of CT images but also ensures that the model can operate effectively in resource-constrained environments, such as poorly equipped hospitals, thereby assisting doctors in the accurate classification of diseases. The core contribution of this study lies in its ability to balance model complexity and resource efficiency, achieving high classification accuracy while ensuring that the model is accessible for practical use in diverse medical settings.
2. Methodology
2.1. Dataset description and preprocessing
This study used a dataset of 44 Classes of Brain Tumor magnetic resonance imaging (MRI) Images in Kaggle [8]. A private collection of T1, contrast-enhanced T1, and T2 magnetic resonance images separated by brain tumor type. A total of 14 cancer images were clearly classified by radiologists. In terms of data processing, this study creates a data enhancement layer before data is input into the model. By way of mirror inversion and adjustment of sharpness. Thus, the number of data sets can be effectively expanded. Then, the data of different diseases in the data set were divided into the training set and the test set according to the ratio of 8:2. The sample is shown in the Figure 1.
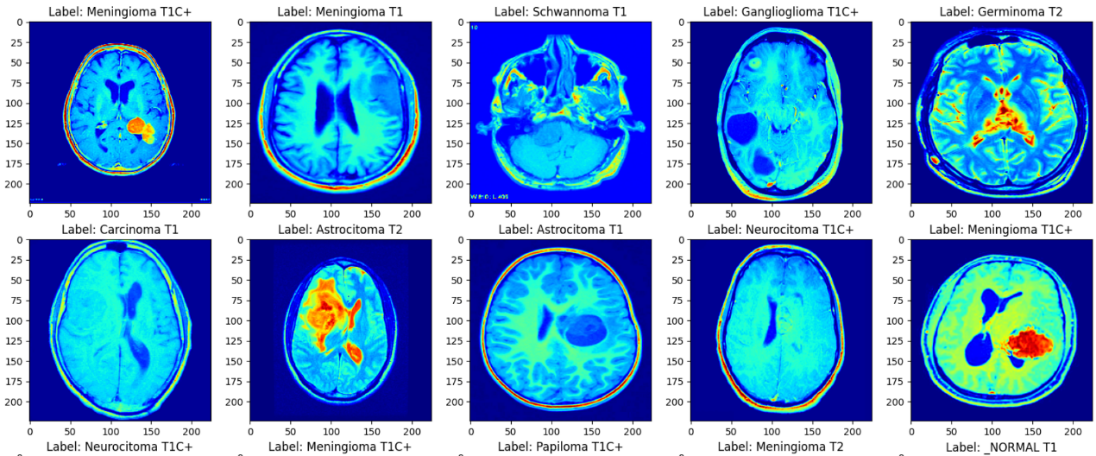
Figure 1. The samples of the MRI images.
2.2. Proposed approach
In this study, author aims to identify a model that minimizes training time while maintaining high accuracy. To achieve this, author systematically compares the training speed and construction complexity of various models. By training the same batch of data on different models and optimizing each one, author assesses their performance to determine which model achieves the best balance of efficiency and accuracy. Through rigorous experimentation and optimization, this study identifies the most practical approach, as illustrated in Figure 2. This method ensures that the chosen model not only delivers high training accuracy but also operates with reduced training time, making it more feasible for real-world applications.
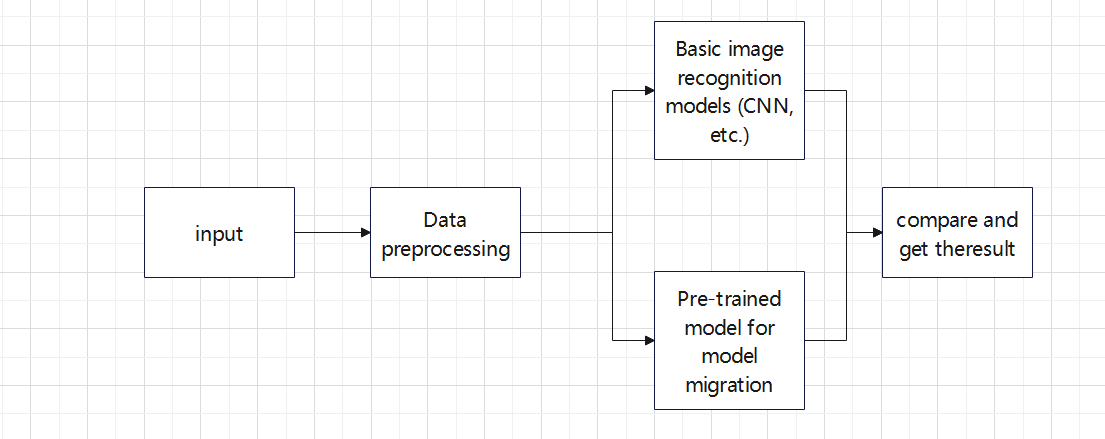
Figure 2. The pipeline of the study.
2.2.1. EfficientNetV2. The basic architecture of EfficientNet is built by CNN, which consists of one or more convolutional layers and a fully connected top layer, as well as associated weights and pooling layers. In the past experience, increasing the number and depth of convolutional nuclei in the network may effectively improve the performance of the network. In order to make the model better applicable to other tasks, increasing the depth of the network is a very good solution. However, too deep a network will cause the gradient to disappear, making training difficult. Therefore, Neural Architecture Search (NAS) is used here to get the most basic structure of the model [9]. As shown in Figure 3, the MBConv structure consists mainly of a 1*1 ordinary convolution (for dimensionality raising), a k*k depthwise Conv convolution, an Squeeze-and-Excitation (SE) module, a 1*1 ordinary convolution (for dimensionality reduction), and a Droupout layer.
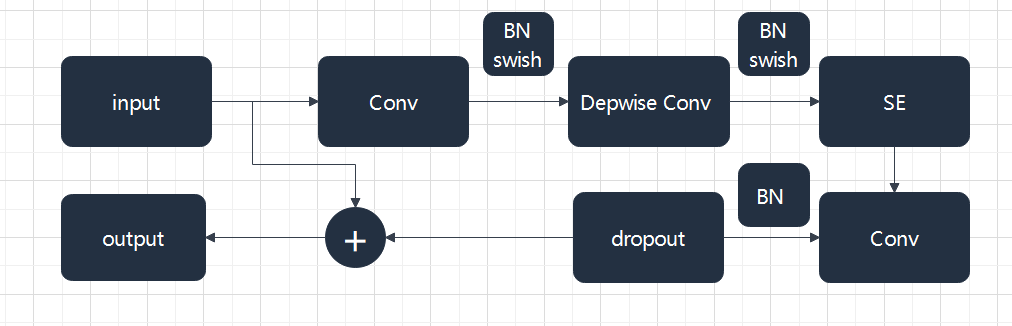
Figure 3. MBConv be built.
EfficientNetV2 resolves several problems with EfficientNetV1. First, it aims at the problem that the training speed is too slow when the training image size is very large. It introduced Fused-MBConv. MBConv and Depthwise Convolution (DWConv) are fused into a 3*3 convolution. Finally, it is found that the training time can be reduced effectively in the early stage of network training. Through a search using NAS technology, it was determined that replacing the first three MBConvs on the network was optimal. Secondly, the depth and width of each stage in V1 are equally enlarged, and the author optimizes it by using the unequal scaling strategy, and finally obtains a better solution by adjusting the parameters of width and depth.
2.2.2. Vision Transformer (ViTs). Vit is a model proposed by Google in 2020 to directly apply transformer to image classification [10] Vit is based on the converter architecture used in natural language processing (NLP), which converts text into sequence tags and generates text embeddings (see in Figure 4). With Vit, input images are preprocessed into tokens, and transfromer calculates the correlation between each token. This is different from the traditional CNN through the convolution and pooling of downward sampling. The transformer encoder block consists of three parts. For layer normalization, apply it to the patch and pay attention to the computation acceleration. Here, for a certain sample, calculate the mean and variance of all feature graphs of the sample, and then normalize the sample. It is more space-efficient than batch normalization, thus saving computing resources. For multi-head attention, it is used to generate and connect attention heads for all patches to capture local and global dependencies in the image. For multilayer Perceptrons (MLPS), as shown on the right side of figure5, it takes the Gaussian Error Linear Unit (GELU) as the activation function, passing the attention head to the two dense layers. The thickness of the input feature layer is expanded four times by a fully connected layer, and then restored to the original number by a fully connected layer, so that the shape of the thick tensor through the Multilayer Perceptron (MLP) module remains unchanged (see in Figure 5).
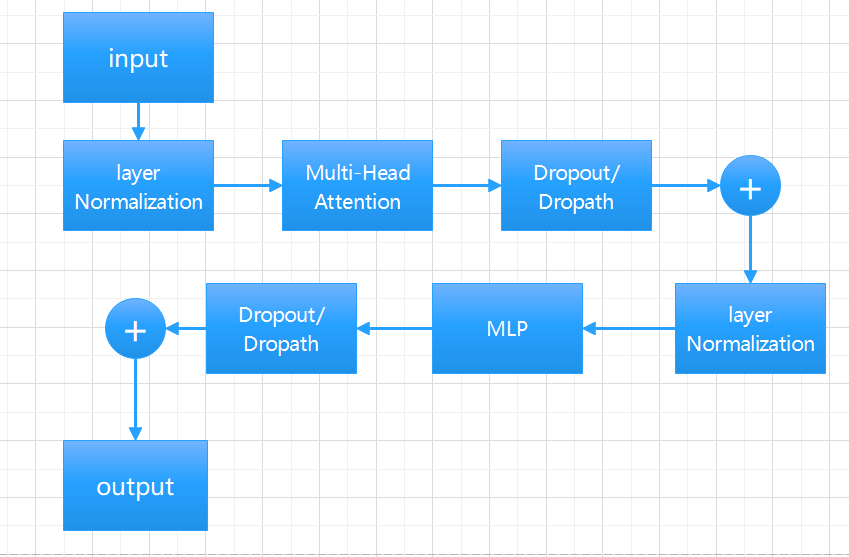
Figure 4. Encoder Block.
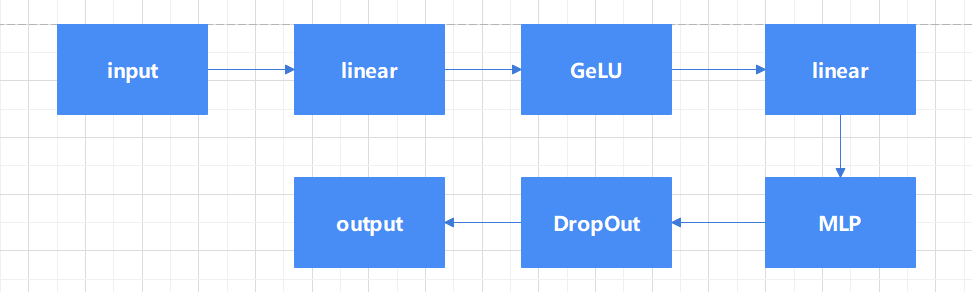
Figure 5. MLP model.
However, since the model has no inductive bias at all, position coding is required, which requires a large data set in the pre-training stage to ensure the model effect. However, the effect of the model after pre-training is very excellent, and it can be used to fine-tune the model directly.
3. Result and Discussion
3.1. Result of Basic CNN
From the Figure 6, it can be seen from the figure that CNN without any modification requires a longer number of iterations and can only be roughly classified, which does not play a good role in disease subdivision. However, it can basically meet the distinction of brain cancer.
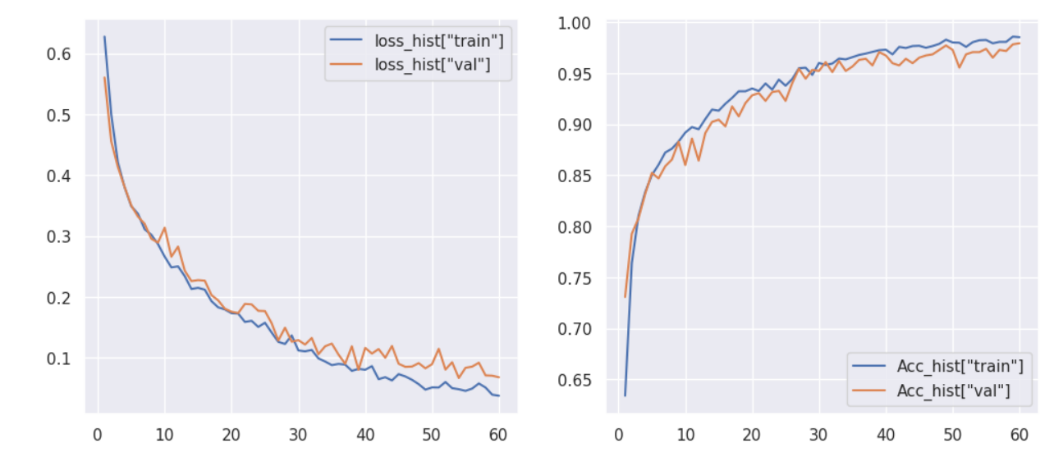
Figure 6. CNN training epochs and accuracy.
3.2. Result of EfficientlyNetV2
From the Figure 7, thesis observe that the model can converge to a training loss that is lower than the validation loss. And no fitting occurred during the training of the model. thesis also note that validation loss convergence is a bit unstable. But compared with the traditional CNN model, it has fewer iterations and higher accuracy. Because using a pre-trained model allows it to match tasks quickly so that it can complete the task in a shorter time.
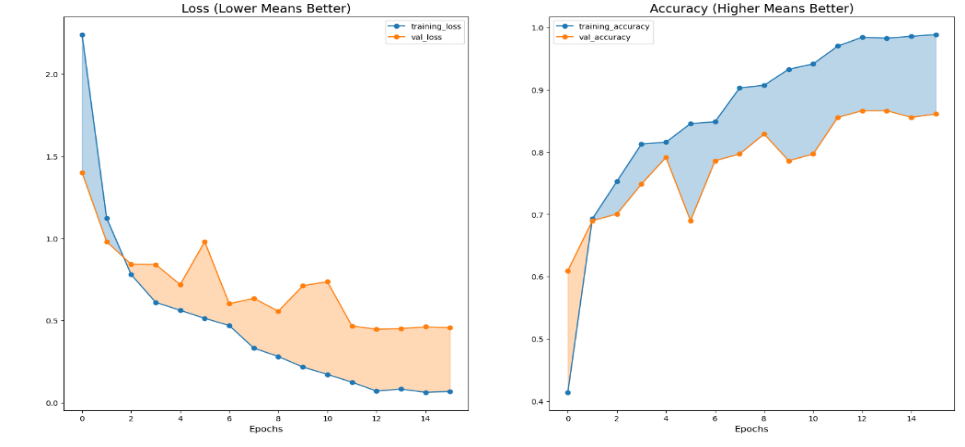
Figure 7. EfficientlyNetV2 training epochs and accuracy.
It can be seen from the table 1 that its prediction accuracy for most diseases is higher than 80%, and only for some diseases with very small sample size (sample size is less than or equal to five), the prediction accuracy is low. It shows that the model can also perform a good task in the subdivision of disease.
3.3. Result of Vision Transformer (ViTs)
Thesis can observe that the training loss in Figure 8 is lower than the validation loss and the gap between the two is smaller than the efficientNet, and the accuracy iteration rate is faster. However, the convergence of validation loss is also unstable. However, there is no overfitting phenomenon, indicating that the model can still guarantee the accuracy on small samples. It can also be found from the table 2 that only when the number of disease samples is extremely small, it will lead to a large deviation in the prediction, and when the number of disease types meets the basic learning needs, it can achieve a high accuracy. This is consistent with what the Google team expected when developing this model.
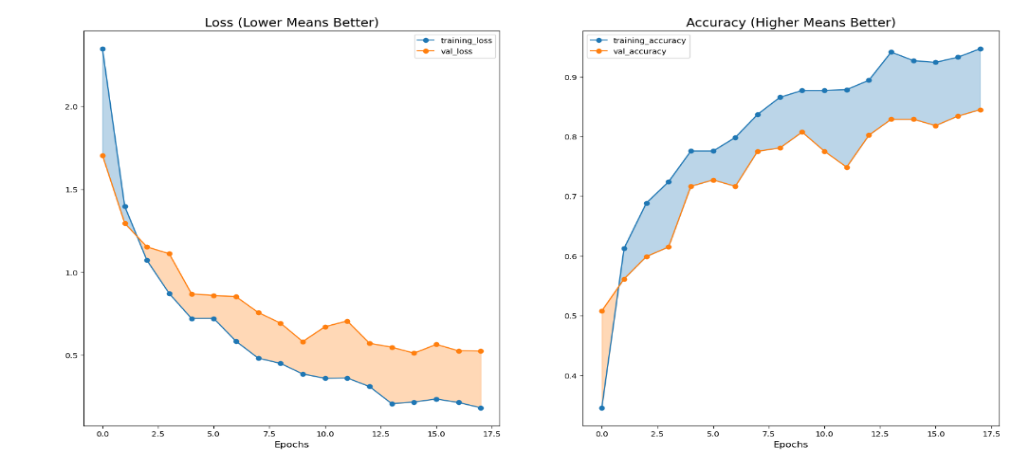
Figure 8. Vision Transformer training epochs and accuracy.
Table 1. Training result about EfficientNetV2.
precision | recall | F1-socre | Support | |
Astrocitoma T1 | 1.00 | 1.00 | 1.00 | 13 |
Astrocitoma T1C+ | 1,00 | 0.93 | 0.96 | 14 |
Astrocitoma T2 | 0.78 | 0.78 | 0.78 | 9 |
Ependimoma T1C+ | 0.50 | 0.50 | 0.50 | 2 |
Ependimoma T2 | 0.43 | 1.00 | 0.60 | 3 |
Ganglioglioma T1 | 1.00 | 1.00 | 1.00 | 3 |
Ganglioglioma T1C+ | 0.67 | 1.00 | 0.80 | 2 |
Ganglioglioma T2 | 0.50 | 1.00 | 0.67 | 1 |
Germinoma T2 | 1.00 | 0.33 | 0.50 | 3 |
Meduloblastoma T2 | 1.00 | 1.00 | 1.00 | 4 |
Meningioma T1 | 1.00 | 0.91 | 0.95 | 44 |
Meningioma T1C+ | 0.94 | 0.98 | 0.96 | 50 |
Meningioma T2 | 0.93 | 0.84 | 0.89 | 32 |
Neurocitoma T1 | 0.73 | 1.00 | 0.84 | 8 |
Neurocitoma T1C+ | 0.95 | 0.90 | 0.92 | 20 |
Neurocitoma T2 | 0.67 | 0.80 | 0.73 | 5 |
Papiloma T1 | 1.00 | 1.00 | 1.00 | 11 |
Papiloma T1C+ | 1.00 | 0.94 | 0.97 | 18 |
Papiloma T2 | 1.00 | 0.67 | 0.80 | 3 |
Schwannoma T1 | 0.75 | 1.00 | 0.86 | 6 |
Schwannoma T1C+ | 1.00 | 0.60 | 0.75 | 5 |
Schwannoma T2 | 1.00 | 0.86 | 0.92 | 7 |
_NORMAL T1 | 0.94 | 1.00 | 0.97 | 15 |
_NORMAL T2 | 0.75 | 1.00 | 0.86 | 3 |
accuracy | 0.91 | 291 | ||
macro avg | 0.86 | 0.88 | 0.84 | 281 |
weighted avg | 0.93 | 0.91 | 0.92 | 281 |
Table 2. Training result about Vision Transformer.
precision | recall | F1-socre | Support | |
Astrocitoma T1 | 1.00 | 1.00 | 1.00 | 13 |
Astrocitoma T1C+ | 1.00 | 0.93 | 0.96 | 14 |
Astrocitoma T2 | 0.78 | 0.78 | 0.78 | 9 |
Ependimoma T1C+ | 0.50 | 0.50 | 0.50 | 2 |
Ependimoma T2 | 0.43 | 1.00 | 0.60 | 3 |
Ganglioglioma T1 | 1.00 | 1.00 | 1.00 | 3 |
Ganglioglioma T1C+ | 0.67 | 1.00 | 0.80 | 2 |
Ganglioglioma T2 | 0.50 | 1.00 | 0.67 | 1 |
Germinoma T2 | 1.00 | 0.33 | 0.60 | 3 |
Meduloblastoma T2 | 1.00 | 1.00 | 1.00 | 4 |
Meningioma T1 | 1.00 | 0.91 | 0.95 | 44 |
Meningioma T1C+ | 0.94 | 0.98 | 0.96 | 50 |
Meningioma T2 | 0.93 | 0.84 | 0.69 | 32 |
Neurocitoma T1 | 0.73 | 1.00 | 0.84 | 8 |
Neurocitoma T1C+ | 0.95 | 0.90 | 0.92 | 20 |
Neurocitoma T2 | 0.67 | 0.80 | 0.73 | 5 |
Papiloma T1 | 1.00 | 1.00 | 1.00 | 11 |
Papiloma T1C+ | 1.00 | 0.94 | 0.97 | 18 |
Papiloma T2 | 1.00 | 0.67 | 0.80 | 3 |
Schwannoma T1 | 0.75 | 1.00 | 0.86 | 6 |
Schwannoma T1C+ | 1.00 | 0.60 | 0.75 | 5 |
Schwannoma T2 | 1.00 | 0.86 | 0.92 | 7 |
_NORMAL T1 | 0.94 | 1.00 | 0.97 | 15 |
_NORMAL T2 | 0.75 | 1.00 | 0.86 | 3 |
accuracy | 0.91 | 281 | ||
macro avg | 0.86 | 0.88 | 0.84 | 281 |
weighted avg | 0.93 | 0.91 | 0.92 | 281 |
4. Conclusion
The study highlights that such models can improve early diagnosis and classification of diseases in primary hospitals, where distinguishing between conditions can be particularly challenging and resources are often scarce. By providing a robust and accurate diagnostic tool, the model helps alleviate the strain on primary doctors who may lack extensive resources. Future work should focus on enhancing these models by incorporating additional training mechanisms and expanding their capabilities. This includes integrating new medical imaging data to continuously refine and improve the models. Additionally, exploring the ability to classify a broader range of diseases could further enhance its utility in various medical contexts. By continually updating and optimizing the model, thesis can support medical staff in delivering more precise and timely diagnoses, ultimately improving patient outcomes.
References
[1]. DeAngelis L M 2001 Brain tumors New England journal of medicine vol 344 no 2 pp 114-123
[2]. Santana L S Diniz J C B Gasparri L M G et al. 2024 Application of Machine Learning for Classification of Brain Tumors: A Systematic Review and Meta-Analysis World Neurosurgery
[3]. Shyamala B Brahmananda S H D 2023 Brain tumor classification using optimized and relief-based feature reduction and regression neural network Biomedical Signal Processing and Control vol 86 p 105279
[4]. Ismael S A A Mohammed A Hefny H 2020 An enhanced deep learning approach for brain cancer MRI images classification using residual networks Artificial intelligence in medicine vol 102 p 101779
[5]. Emam M M Samee N A Jamjoom M M et al. 2023 Optimized deep learning architecture for brain tumor classification using improved Hunger Games Search Algorithm Computers in Biology and Medicine vol 160 p 106966
[6]. Tejashwini P S Thriveni J Venugopal K R 2024 EBT Deep Net: Ensemble brain tumor Deep Net for multi-classification of brain tumor in MR images Biomedical Signal Processing and Control vol 95 p 106312
[7]. Rajeev S K Rajasekaran M P Vishnuvarthanan G et al. 2022 A biologically-inspired hybrid deep learning approach for brain tumor classification from magnetic resonance imaging using improved gabor wavelet transform and Elmann-BiLSTM network Biomedical Signal Processing and Control vol 78 p 103949
[8]. Tan M Le Q 2021 Efficientnetv2: Smaller models and faster training International conference on machine learning PMLR pp 10096-10106
[9]. Tan M 2019 Efficientnet: Rethinking model scaling for convolutional neural networks Preprint: arXiv:1905.11946
[10]. Dosovitskiy A 2010 An image is worth 16x16 words: Transformers for image recognition at scale Preprint: arXiv:2010.11929
Cite this article
Zhao,C. (2024). Efficient Brain Cancer Classification Based on Advanced CNN and Transformer Technologies. Applied and Computational Engineering,103,86-94.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 2nd International Conference on Machine Learning and Automation
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. DeAngelis L M 2001 Brain tumors New England journal of medicine vol 344 no 2 pp 114-123
[2]. Santana L S Diniz J C B Gasparri L M G et al. 2024 Application of Machine Learning for Classification of Brain Tumors: A Systematic Review and Meta-Analysis World Neurosurgery
[3]. Shyamala B Brahmananda S H D 2023 Brain tumor classification using optimized and relief-based feature reduction and regression neural network Biomedical Signal Processing and Control vol 86 p 105279
[4]. Ismael S A A Mohammed A Hefny H 2020 An enhanced deep learning approach for brain cancer MRI images classification using residual networks Artificial intelligence in medicine vol 102 p 101779
[5]. Emam M M Samee N A Jamjoom M M et al. 2023 Optimized deep learning architecture for brain tumor classification using improved Hunger Games Search Algorithm Computers in Biology and Medicine vol 160 p 106966
[6]. Tejashwini P S Thriveni J Venugopal K R 2024 EBT Deep Net: Ensemble brain tumor Deep Net for multi-classification of brain tumor in MR images Biomedical Signal Processing and Control vol 95 p 106312
[7]. Rajeev S K Rajasekaran M P Vishnuvarthanan G et al. 2022 A biologically-inspired hybrid deep learning approach for brain tumor classification from magnetic resonance imaging using improved gabor wavelet transform and Elmann-BiLSTM network Biomedical Signal Processing and Control vol 78 p 103949
[8]. Tan M Le Q 2021 Efficientnetv2: Smaller models and faster training International conference on machine learning PMLR pp 10096-10106
[9]. Tan M 2019 Efficientnet: Rethinking model scaling for convolutional neural networks Preprint: arXiv:1905.11946
[10]. Dosovitskiy A 2010 An image is worth 16x16 words: Transformers for image recognition at scale Preprint: arXiv:2010.11929









