1. Introduction
Drug addiction is now understood as a chronic, relapsing brain disease. Researchers characterize it by compulsive drug-seeking behaviors despite adverse consequences [1]. Neuroscience models show that addiction involves multiple overlapping neural circuits, such as reward, memory, cognitive control, and salience. Chronic substance use renders them dysregulated. For example, Volkow’s model highlights dysfunction in cortico-striatal-limbic loops. Addictive substances dysregulate neural reward circuits and compromise prefrontal control [2]. Empirical resting-state fMRI studies in addicted populations (e.g. chronic heroin users) have found altered functional connectivity in these circuits. Meta-analyses of fMRI studies in cocaine, cannabis, alcohol, and nicotine users have found common functional alterations in dorsal striatal and prefrontal circuits during cognitive and reward-related tasks [2]. Such evidence indicates that addiction reshapes large-scale brain network organization, amplifying drug-salient pathways and degrading executive-control networks.
Advanced neuroimaging analyses have adopted network science approaches to capture these changes. The brain can be modeled as a graph whose nodes are anatomically-defined regions and whose edges reflect functional or structural connections. This graph formulation enables use of machine learning on non-Euclidean data. Recently, graph neural networks (GNNs), deep learning models that operate on graph-structured data, have emerged as powerful tools for brain connectivity analysis [3]. GNNs propagate information along graph edges and learn hierarchical node embeddings, allowing them to capture complex interaction patterns in brain networks. For instance, attention-based GNNs can assign higher weights to the most important connections, improving interpretability. In brain imaging, GNNs have been utilized across a wide range of disorders, providing state-of-the-art classification and feature selection by leveraging the full graph topology [3, 4].
In this paper, the FGSAN framework has been proposed to identify addiction-related biomarkers from fMRI-derived brain graphs. FGSAN (and related models) combine graph attention mechanisms with a Bayesian feature selector and a classifier to detect altered connectivity patterns in addiction. This paper covers the neuroscience of addiction-related connectivity and the theoretical underpinnings of FGSAN, and analyzes key case studies where GNNs – including FGSAN-like models – have been used to classify addiction and related disorders.
2. Theoretical foundations
2.1. Addiction-related brain connectivity
Drug addiction reflects comprehensive alterations in brain networks that related to motivation, reward learning, and control. According to animal and human studies, chronic substance exposure will produce long-lasting synaptic and structural alterations in the reward circuit, including plasticity in the nucleus accumbens, ventral tegmental area, and prefrontal cortex [1, 2]. These neuroadaptations manifest as altered patterns of functional connectivity during rest and task states. For example, a meta-analysis of task-based fMRI studies shows that substance use disorders are associated with common functional alterations in frontostriatal circuits engaged in reward processing and executive control [2].
These alterations align with the idea that addiction amplifies the salience of drug-related cues while diminishing cognitive control. Several major neural circuits are critically involved: the reward circuit, comprising the nucleus accumbens, ventral pallidum, and ventral tegmental area, underlies reinforcement processes; the amygdala and hippocampus form a memory–learning circuit that encodes drug-associated cues; the dorsolateral prefrontal cortex and dorsal anterior cingulate cortex constitute a cognitive control network essential for inhibitory regulation; and the orbitofrontal cortex contributes to a motivation–salience circuit that assigns value to stimuli. The progression to compulsive use is thought to be driven by dysregulations in motivational circuits, particularly frontostriatal systems involved in reward processing, habit formation, and executive control [2]. The net effect is a functional network that is hypersensitive to drug-related inputs but relatively deficient in executive regulation.
FMRI provides a window into these network changes. Functional neuroimaging, including fMRI, is a powerful tool for identifying alterations in brain circuits, with meta-analyses providing robust evidence for common changes across substance use disorders [2]. Studies in addiction have leveraged rs-fMRI to identify biomarkers: for example, altered local coherence (ReHo) and connectivity in prefrontal, temporal, and visual regions have been reported in internet gaming disorder (a behavioral addiction) [5]. Chinese neuroimaging reviews similarly emphasize that drug and behavioral addiction involve widespread brain network alterations [5, 6]. Overall, both English and Chinese literature converge on the view that drug addiction is a chronic brain disorder with systematic connectivity disruptions [1, 6]. This motivates analysis methods that can integrate full-brain connectivity patterns to distinguish addicted from non-addicted subjects and pinpoint affected circuits. Functional connectivity differences between remission and ongoing addiction is shown in Figure 1.
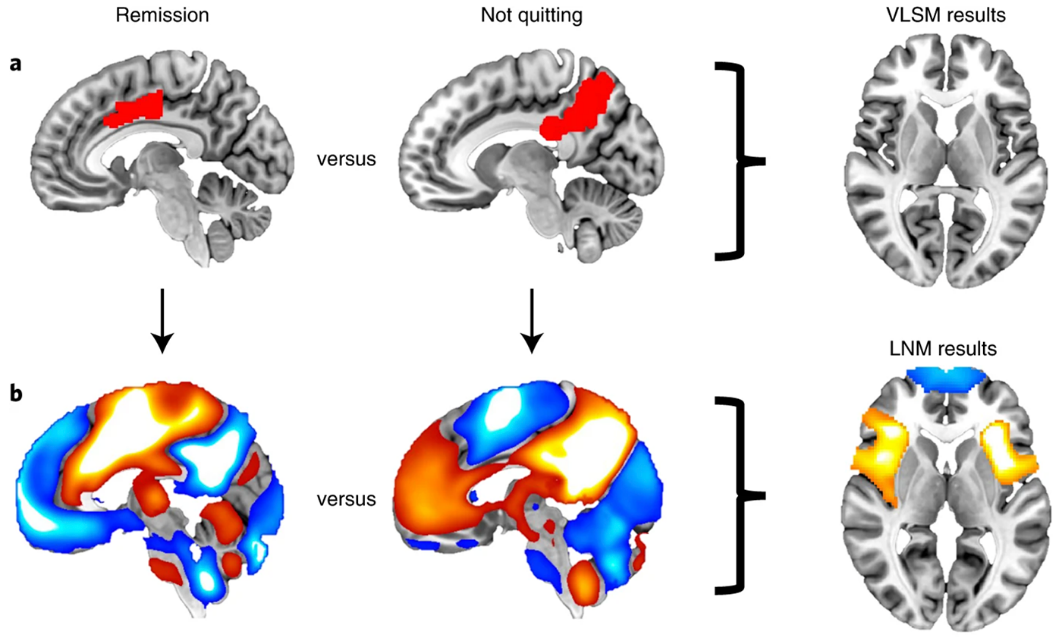
2.2. Feature-selected graph spatial attention networks
Graph neural networks for brain analysis typically consist of a graph encoder (to learn embeddings), a feature selection component, and a classifier. The FGSAN approach combines a graph spatial attention encoder with a Bayesian feature selector and a downstream MLP classifier. Below we outline each component.
2.2.1. Graph spatial attention encoder
The encoder component constructs meaningful embeddings of brain-network graphs using an attention mechanism. In FGSAN, the fMRI-derived brain graph (nodes = brain regions, edges = functional connections) is input to a multi-layer graph attention network. Unlike standard graph convolution, graph attention networks compute attention coefficients for each edge, allowing the model to weight neighbor contributions adaptively. In particular, FGSAN uses spatial attention, meaning that it integrates spatial information about regions (e.g. anatomical coordinates or predefined order) to inform attention weights [8]. Concretely, each graph layer computes where
In the above equation, denotes the feature vector of node
2.2.2. Bayesian feature selector
To prevent overfitting and focus on discriminative brain connections, FGSAN includes a Bayesian feature selection (BFS) module between the encoder and the classifier. BFS introduces a probabilistic mask that multiplies node features or edge features by binary random variables, enforcing sparsity. During training, the model learns probabilities for each feature being “selected.” In practice, a feature is kept with some probability (and dropped otherwise), forming a variational dropout that performs feature selection. Gong et al. showed that adding BFS significantly improved performance, for example, raising accuracy from 75.5% to 81.3% on a nicotine addiction task [9]. The BFS acts as a structural regularizer: it “restricts” the model to use only the most relevant nodes and edges. Intuitively, BFS encourages the model to ignore noisy or redundant connections, enabling it to identify a sparse subset of biomarkers [8, 9]. This improves generalization and interpretability, since the selected features correspond to the most informative brain regions.
2.2.3. MLP classifier
After encoding and feature selection, the reduced feature representation is fed to a simple multi-layer perceptron (MLP) for classification. In FGSAN-style models, the final graph-level feature is concatenated into a vector which an MLP uses to predict labels. Gong et al. note that “the selected and readout features are delivered to a MLP to derive the final identification”. The MLP learns to map the high-level graph embedding into diagnostic categories. The combined model (attention encoder + BFS + MLP) is trained end-to-end using standard supervised loss [9].
In summary, FGSAN integrates graph attention, Bayesian feature selection, and an MLP. This architecture is motivated by the need to both exploit the non-Euclidean graph structure of brain data and to discover a small set of neural biomarkers.
3. Case discussion and analysis
3.1. Feature-selected graph spatial attention network (FGSAN) for addiction
Gong et al. introduced the FGSAN model to identify addiction-related brain networks in a rat fMRI dataset [8]. They construct each brain scan as a weighted graph (150 nodes for rat regions, edges by Pearson correlation). The graph spatial attention encoder consists of multiple graph-attention layers that incorporate each region’s spatial attributes. According to the authors, “a graph spatial attention encoder is employed to capture the features of spatiotemporal brain networks with spatial information”. In effect, each layer learns attention coefficients that weigh nearby regions’ influence, yielding embeddings sensitive to anatomical organization. The spatial attention is key: it allows the model to focus on region pairs that exhibit addiction-related coupling changes.
Simultaneously, the model applies a Bayesian feature selection step. During training, each node’s feature (or each graph-level statistic) is multiplied by a learned Bernoulli mask, effectively selecting a subset of nodes. This strongly regularizes the model. Gong et al. report that using BFS “optimizes the model and improves classification” [8]. In their experiments, FGSAN achieved significantly higher accuracy than baseline GNNs. Although exact numbers are not reported in the abstract, their ablation on a successor model (SARN) shows that an analogous setup yields 81.3% accuracy with BFS versus 75.5% without [9].
Finally, selected graph features are fed to an MLP for graph classification. The model was trained end-to-end in PyTorch, using stochastic gradient descent. Gong et al. found that FGSAN not only classifies addiction vs. control effectively, but also highlights biologically plausible biomarkers. Their paper lists the top five brain regions identified by FGSAN: midbrain, diagonal domain, primary motor cortex, hippocampal formation, and insular cortex [9]. These regions are known to be involved in dopaminergic reward processing and interoception, consistent with nicotine addiction mechanisms. In sum, FGSAN represents a powerful integration of attention and feature selection for graph-level analysis of addiction.
3.2. Spatial attention recurrent network (SARN) with BFS
An extension of FGSAN, Gong et al. proposed SARN to incorporate temporal dynamics along with spatial attention. Using the same rat fMRI data, they segment each 800-time-point series into four sequential windows, constructing a dynamic graph sequence. The SARN encoder stacks three graph spatial-attention layers (as in FGSAN) followed by a specialized attention-based recurrent layer. This design allows the model to capture both static spatial connectivity and evolving temporal patterns. Indeed, the SARN architecture is “composed of three graph spatial attention layers and one sliding-window attention recurrent layer” [9]. The sliding-window RNN attends over the sequence of graph embeddings, learning how connectivity features change over time.
The Bayesian feature selector from FGSAN is retained. As before, BFS sparsifies the node embeddings before classification. Gong et al. explicitly states that in SARN, the BFS “strategy is adopted to optimize the model and improve classification tasks by restricting features”.
In performance tests, SARN markedly outperformed simpler models. An ablation study shows that replacing SARN’s encoder with a vanilla Graph Attention Network (GAT) yielded only 67.8% accuracy, whereas the full SARN achieved 75.5%. When BFS was added, accuracy rose to 81.3%. Thus, including temporal information in the graph encoder significantly improved discrimination (from ~68% to ~75%), and BFS provided an additional boost to ~81%. The authors explain this by noting that spatial attention adds positional context (important brain map geometry), while the recurrent layer captures time-dependent dynamics. The BFS then selects the most task-relevant nodes, further enhancing classification [9].
Crucially, SARN maintained FGSAN’s interpretability benefits. Gong et al. visualized the top-weighted regions (e.g. midbrain, hippocampus) and confirmed they align with known addiction circuits. They also compared SARN+BFS to contrastive GNN methods and found SARN superior on all metrics [9]. In summary, the SARN case demonstrates that combining graph spatial attention with temporal recurrence and Bayesian selection yields strong, interpretable models for dynamic addiction networks.
3.3. Graph Diffusion Reconstruction Network (GDRN)
While FGSAN/SARN are discriminative models, Jing et al. introduced a generative graph model for addiction called the Graph Diffusion Reconstruction Network (GDRN) [10]. GDRN aims to capture addiction-related connectivity by learning a diffusion-based reconstruction of brain graphs. In this framework, the model is trained to generate synthetic graph samples that match the distribution of real addiction-network graphs. A key component is a diffusion reconstruction module: the network takes a latent graph representation and iteratively “diffuses” it through a learned process, reconstructing adjacency patterns. This diffusion mechanism ensures that the generated graphs maintain the overall data distribution of the training set.
On the nicotine-addiction rat fMRI dataset, GDRN demonstrated its ability to reproduce the connectivity patterns of addicted brains. Experiments reported that GDRN effectively “captures addiction-related brain connectivity” and that the diffusion module “enhanced the ability to reconstruct nicotine addiction-related brain networks” [10]. In other words, by training the model to reconstruct graphs rather than only classify them, the authors ensure the latent space encodes the essential structure of addiction circuitry. Although quantitative metrics for GDRN are not detailed in the abstract, the reported results claim successful exploration of addiction networks. This case illustrates an alternative perspective: a generative GNN can model the distribution of addicted brain graphs and thus highlight common connectivity features of addiction. It complements the FGSAN/SARN cases by providing a holistic reconstruction-based approach.
3.4. GNN classification in schizophrenia (similar neuropsychiatric domain)
To contextualize how GNN methods generalize to other brain disorders, we consider Sunil et al., who used a deep graph convolutional neural network (DGCNN) on human resting-state fMRI to classify schizophrenia [4]. Although not an addiction study, their methodology mirrors FGSAN’s spirit: they represent each subject’s brain as a graph (nodes = regions, edges = functional correlations) and apply GNNs for diagnosis. Their DGCNN achieved an average accuracy of 0.82 (AUC ≈ 0.84) in distinguishing schizophrenia patients from controls, comparable to classical machine-learning models. Importantly, they also incorporated feature selection to identify potential network biomarkers. The study underscores that GNNs can uncover distributed connectivity signatures of brain disorders: in this case, schizophrenia-related patterns, while our primary focus is on addiction [4]. This cross-domain example shows that graph-based deep learning techniques are broadly applicable to neurological and psychiatric disorders, and techniques like spatial attention or BFS could similarly enhance such models.
4. Conclusion
Recent work demonstrates that graph neural networks – especially those augmented with attention and feature selection – are effective for analyzing addiction-related brain connectivity. Key trends include the use of attention mechanisms to weight important neural pathways and Bayesian selectors to prune irrelevant signals. Compared to conventional methods, these GNN models automatically leverage the non-Euclidean structure of brain networks, learning end-to-end features that yield high classification accuracy. For instance, spatial-attention layers capture relationships between brain regions, while recurrent layers model temporal dynamics. Constraining the model with a BFS procedure, it will help focus learning on features relevant for addiction. Models constructed in this way often perform well in experimental settings, such as frequently exceeding 80% accuracy. Moreover, they also highlight interpretable biomarkers such as the midbrain, hippocampus, and insula that are consistent with neuroscience findings. Nonetheless, important challenges still remain. Training datasets are typically small, animal cohorts or limited human samples, which increases the risk of overfitting. Combined with differences across species, scanner types, and preprocessing pipelines, the high dimensionality and heterogeneity of fMRI-derived graphs further reduce model generalization. Finally, although attention mechanisms offer some interpretability, complementary explainable-AI techniques are still required to fully explain how these models make decisions. Finally, computational complexity of GNNs can be high on large brain graphs, requiring efficient algorithms for scalability.
In conclusion, FGSAN-like models represent a promising direction for brain network analysis in addiction. They utilize graph theory, deep learning, and Bayesian feature selection to reveal the altered circuitry of addiction. Future work needs to validate these approaches in larger human cohorts and testing their applicability to other neuropsychiatric disorders. Important directions include integrating multi-modal imaging, designing dynamic GNN architectures that capture evolving connectivity patterns, and using transfer learning to improve robustness across studies and scanners. Ultimately, graph neural networks provide a promising framework for discovering network-level biomarkers of addiction. What’s more, they could support the development of connectivity-informed diagnostics and interventions.
References
[1]. Parvaz, M. A., Moeller, S. J., & Goldstein, R. Z. (2024). Neuroimaging biomarkers in addiction. medRxiv.
[2]. Schulze, L., et al. (2020). Common and separable neural alterations in substance use disorders: A coordinate-based meta-analysis of functional neuroimaging studies in humans. Human Brain Mapping, 41(16), 4459–4477.
[3]. Mohammadi, H., & Karwowski, W. (2025). Graph neural networks in brain connectivity studies: Methods, challenges, and future directions. Brain Sciences, 15(1), 17.
[4]. Sunil, G., Gowtham, S., Bose, A., Harish, S., & Srinivasa, G. (2024). Graph neural network and machine learning analysis of functional neuroimaging for understanding schizophrenia. BMC Neuroscience, 25, Article 2.
[5]. Chen, J., Zhang, Y., Niu, X., Zhang, M., Ma, L., & Cheng, J. (2024). A resting-state functional magnetic resonance imaging study of brain functional abnormalities in Internet gaming disorder patients. Magnetic Resonance Imaging (CJMRI), 15(8), 59–64.
[6]. Zheng, Y., Zhai, T., Lin, X., et al. (2024). The resting-state brain activity signatures for addictive disorders. Med, 5(3).
[7]. Google. (2025, August 23). Brain connectivity and addiction fMRI. Google. [Online image]. hotp-6-2022-full.png
[8]. Gong, C., Jing, C., Pan, J., & Wang, S. (2022). Feature-selected graph spatial attention network for addictive brain-networks identification. arXiv preprint arXiv: 2207.00583.
[9]. Gong, C., Chen, X., Mughal, B., & Wang, S., et al. (2023). Addictive brain-network identification by spatial attention recurrent network with feature selection. Brain Informatics, 10(1), Article 2.
[10]. Jing, C., Gong, C., Chen, Z., & Wang, S. (2023). Graph diffusion reconstruction network for addictive brain-networks identification. Brain Informatics. 3974, 133–145.
Cite this article
Wu,Y. (2025). Graph Neural Network Analysis of Addiction-Related Brain Networks. Applied and Computational Engineering,186,25-31.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of CONF-FMCE 2025 Symposium: Semantic Communication for Media Compression and Transmission
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Parvaz, M. A., Moeller, S. J., & Goldstein, R. Z. (2024). Neuroimaging biomarkers in addiction. medRxiv.
[2]. Schulze, L., et al. (2020). Common and separable neural alterations in substance use disorders: A coordinate-based meta-analysis of functional neuroimaging studies in humans. Human Brain Mapping, 41(16), 4459–4477.
[3]. Mohammadi, H., & Karwowski, W. (2025). Graph neural networks in brain connectivity studies: Methods, challenges, and future directions. Brain Sciences, 15(1), 17.
[4]. Sunil, G., Gowtham, S., Bose, A., Harish, S., & Srinivasa, G. (2024). Graph neural network and machine learning analysis of functional neuroimaging for understanding schizophrenia. BMC Neuroscience, 25, Article 2.
[5]. Chen, J., Zhang, Y., Niu, X., Zhang, M., Ma, L., & Cheng, J. (2024). A resting-state functional magnetic resonance imaging study of brain functional abnormalities in Internet gaming disorder patients. Magnetic Resonance Imaging (CJMRI), 15(8), 59–64.
[6]. Zheng, Y., Zhai, T., Lin, X., et al. (2024). The resting-state brain activity signatures for addictive disorders. Med, 5(3).
[7]. Google. (2025, August 23). Brain connectivity and addiction fMRI. Google. [Online image]. hotp-6-2022-full.png
[8]. Gong, C., Jing, C., Pan, J., & Wang, S. (2022). Feature-selected graph spatial attention network for addictive brain-networks identification. arXiv preprint arXiv: 2207.00583.
[9]. Gong, C., Chen, X., Mughal, B., & Wang, S., et al. (2023). Addictive brain-network identification by spatial attention recurrent network with feature selection. Brain Informatics, 10(1), Article 2.
[10]. Jing, C., Gong, C., Chen, Z., & Wang, S. (2023). Graph diffusion reconstruction network for addictive brain-networks identification. Brain Informatics. 3974, 133–145.









