1 Introduction
Graph neural networks (GNNs) [1, 2] attract a lot of attention in representation learning for graph structured data. Transfer learning has become a cornerstone of machine learning advances, especially in optimizing GNNs that specialize in processing graph-structured data [3]. This technology reuses a model customized for one task as the basis for another, effectively solving the challenge of data scarcity and enhancing model performance. Academic research has confirmed that transfer learning can significantly improve the efficacy of GNNs in various tasks [4,5], including but not limited to node and graph classification. This study aims to delve into the application of transfer learning in GNNs to determine its impact on model performance, especially when adapted to novel but relevant datasets [1,6]. The initiative is inspired by the precedent success of transfer learning in different scientific fields, which shows its potential to make GNNs more computationally efficient and versatile in various scenarios.
1.1 Challenges
Leveraging transfer learning to enhance the efficacy of GNNs presents a subtle set of challenges [4]. The most important is the risk of negative transfer, an adverse outcome in which a GNN exhibits performance degradation on a new task due to differences between the new task data and the data on which the model was initially trained [1,7]. This inconsistency can cause the transferred knowledge to backfire and hinder the task. To establish a robust transfer learning framework in GNN, the following key challenges must be solved: first, the occurrence of negative transfer must be reduced [5]; second, the relevance and applicability of the transferred knowledge must be ensured, especially when the task data is different from the training data. Third, there is an obligation to maintain the integrity and validity of the transfer learning process in the context of GNN. In summary, these challenges form the key to refining the transfer learning paradigm to realize its full potential to benefit GNN applications.
1.2 Contributions
To summarize, this work makes the following major contributions: This research is critical to advancing the field of GNN through transfer learning methods. The first contribution is a systematic and extensive exploration of GNN pre-training strategies. The basis for this exploration was the creation of two large-scale, domain-specific pre-training corpora: a chemistry domain dataset containing two million figures and a biology domain dataset containing 395,000 figures [1,2,3]. Such a broad dataset is expected to yield important insights into the impact of domain-specific data volume on the effectiveness of GNN pre-training. The second contribution entails formulating and empirically validating a novel pre-training strategy carefully designed to optimize GNNs for enhanced generalization in out-of-distribution environments [2,4]. This is of particular importance in complex transfer learning scenarios, where flexibility and adaptability across different tasks are crucial. Collectively, these contributions are expected to profoundly impact GNNs’ generalizability, facilitate their application in a wide range of tasks, and potentially facilitate breakthroughs in how GNNs can adapt and perform in a variety of transfer learning environments.
2 Related work
In recent years, transfer learning has become an important method to enhance the capabilities of GNN, which perform well in processing graph-structured data. Well-known studies demonstrate the success of this approach. For example, Koo Verjee et al. demonstrated significant improvements in node and graph classification tasks across various datasets, highlighting the adaptability of transfer learning to different data types. Han et al. proposed an innovative adaptive transfer learning paradigm that leverages self-supervised tasks to refine graph representations, thus extending the practical applications of GNNs. Likewise, recent contributions have expanded the field of transfer learning into new areas. The work of Mari et al. explored the integration of transfer learning with hybrid classical-quantum neural networks, opening up new possibilities for high-resolution image classification. Another study by Levie et al. studied the transferability of spectral graph convolutional networks, providing insights into the efficient transfer of spectral filters on different graphs. Building on these foundational works, this article introduces two additional vital contributions to the recent literature. The study by Yang et al. introduced PTGB, a pretraining framework designed for GNNs targeting brain network analysis, which addresses the scarcity of labeled data in the field by leveraging unsupervised pretraining techniques tailored to the field’s unique needs. Another contribution by Wu et al. proposed a comprehensive strategy to retrain GNNs across more diverse graph domains and evaluate the effectiveness of these pre-trained models under various conditions to gain a broader understanding of the impact of pretraining strategies on GNN generalization capabilities. Collectively, these studies highlight the growing importance of transfer learning in improving the efficiency and scope of graph-based data analysis and highlight ongoing challenges, such as negative transfer when source and target datasets differ.
3 Methods
Our project can be divided into two parts: the first part is a replication study of the previous paper[2, 8] which proposed two node-level pre-training strategies and a graph-level pre-training strategy and combined them together to give the best pre-training strategy on graph neural networks; the second part is to evaluate if the pre-training strategy it proposed can be generalized to broader domains, since this previous work only tested it on biology and chemistry domain. By evaluating the pre-training strategy on broader domains, we can prove the generalization and robustness of the pre-training strategy and the overall knowledge transfer from the source domain to the target domain in graph neural networks.
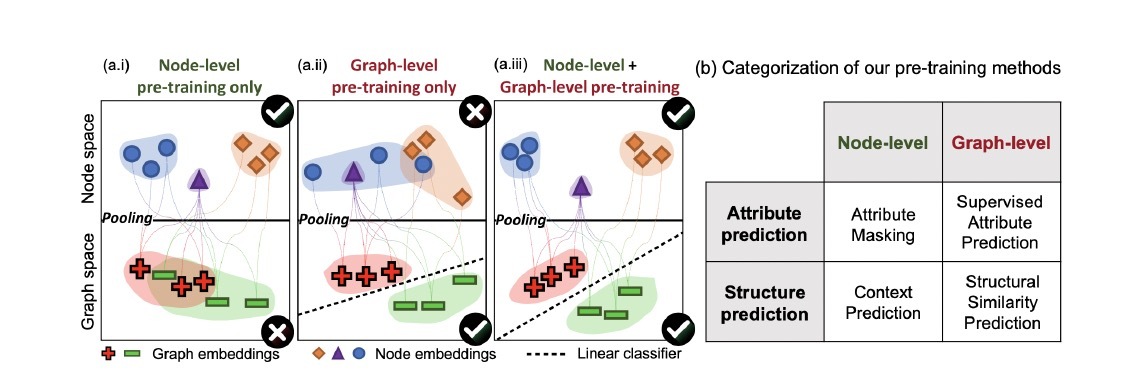
Figure 1. The overview of combining node-level and graph-level pre-training strategy: (a.i) Node-level pretraining only, node embeddings are not composable, and thus resulting graph embeddings are not separable. (a.ii) Graph-level pre-training only, node embeddings do not capture their domain-specific semantics. (a.iii) The pre-training strategy of the original work. Node embeddings are such that nodes of different types are well separated, while the embedding space is also composable.
There are two main categories of pre-training strategies for graph neural networks: node-level and graph-level pre-training. Figure 1 gives an overview of these two strategies and the combining strategy. For node-level pre-training, the paper proposes two methods: Attribute Masking and Context Prediction. Attribute Masking involves randomly masking node or edge attributes and training the GNN to predict those masked attributes based on the neighboring graph structure. Context Prediction uses the node embedding to predict the context structure surrounding each node’s neighborhoods. For graph-level pre-training, the paper uses supervised pre-training on the task of predicting various graph-level attributes. This allows the GNN to learn useful graph-level representations. By combining both the node-level and graph-level pre-training strategies, the model learns useful local node embeddings that capture domain-specific semantics and then further obtains meaningful graph-level representations.
3.1 Node-level pre-training
Figure 2 illustrates the two node-level self-supervised pre-training methods proposed in the previous paper, including Context Prediction and Attribute Masking. Both methods are to force GNN to learn useful local structural and semantic information by predicting the context or attributes. This allows the model to learn meaningful node-level representations.
3.1.1 Context prediction
Context Prediction uses the node embeddings learned by GNN to predict the surrounding graph structure for each node. Specifically, a subgraph centered around a particular node is selected. This subgraph contains the K-hop neighborhood of the center node, where K is the number of GNN layers. The context graph is defined as the graph structure that is between r1 and r2 hops away from the center node. By training to encode the context graph, the model can predict this context embedding using the embedding of the center node.
3.1.2 Attribute masking
Attribute Masking randomly masks node or edge attributes and trains GNN to predict those attributes. First, certain node attributes in the input graph (e.g., atom types) are randomly masked. Instead of training on original graph, GNN uses this masked graph to compute node embeddings. Then model uses these embeddings to predict the masked node/edge attributes.
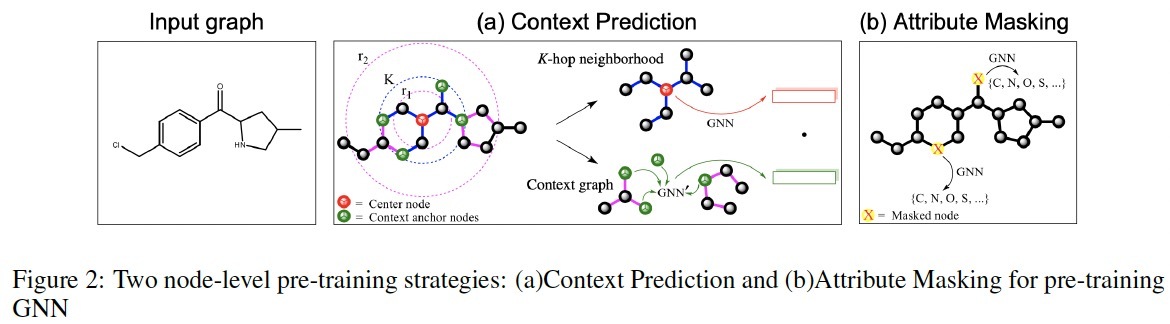
Figure 2. Two node-level pre-training strategies: (a)Context Prediction and (b)Attribute Masking for pre-training GNN
Table 1. Model performance of reproducing
Strategy |
BACE |
BBBP |
Clintox |
|||
AUC |
Precision |
AUC |
Precision |
AUC |
Precision |
|
Without Pre- training |
70.0 |
65.4 |
71.1 |
59.4 |
57.6 |
46.9 |
contextpred |
78.3 |
79.0 |
71.4 |
57.2 |
70.5 |
64.2 |
supervised contextpred |
81.4 |
77.1 |
69.6 |
58.8 |
71.9 |
60.3 |
infomax |
73.1 |
75.0 |
69.5 |
57.6 |
70.9 |
60.1 |
supervised infomax |
78.6 |
80.3 |
67.9 |
60.8 |
72.2 |
56.8 |
edgepred |
81.6 |
78.6 |
70.9 |
58.2 |
67.2 |
46.9 |
supervised edgepred |
78.9 |
73.3 |
71.7 |
61.8 |
71.7 |
58.2 |
masking |
78.7 |
73.5 |
71.3 |
59.2 |
79.9 |
64.5 |
supervised masking |
79.2 |
76.7 |
69.1 |
60.8 |
76.9 |
55.4 |
3.2 Graph-level pre-training
A graph-level pre-training strategy has also been proposed in this paper to learn useful graph-level representations. By using supervised pre-training tasks that predict properties or attributes of entire graphs. For example, in molecular property prediction, GNN can be pre-trained to predict various experimentally measured properties of molecules such as toxicity or binding affinity. By learning to predict these graph-level properties, the model can capture important domain-specific knowledge in its graph-level representations. These pre-trained graph-level embeddings can then be fine-tuned for specific downstream tasks, leading to improved performance.
4 Results & Discussion
4.1 Datasets
By evaluating the node-level pre-training strategies on diverse datasets, we aim to demonstrate the effectiveness and generalizability of the approach within and beyond the domains explored in the original paper. Our experiments cover more graph types and different GNN architectures to provide a comprehensive assessment of the pre-training strategies in a broader context.
4.1.1 Datasets for reproducing
In our replication study, we choose three datasets from MoleculeNet[9], a benchmark for molecular property prediction. MoleculeNet includes multiple public datasets and establishes a standardized evaluation platform for molecular machine learning methods. From MoleculeNet, we select the BACE, BBBP, and ClinTox datasets. BACE is a binary classification task for predicting whether a molecule inhibits the human enzyme associated with Alzheimer’s disease. BBBP is another binary classification task that predicts a molecule’s ability to penetrate the blood brain barrier. ClinTox contains two binary classification tasks related to clinical trial toxicity and FDA approval status.
Table 2. Model perfomance on various datasets
Strategy |
Cora |
Cora (GIN) |
Cora & PubMed |
Amazon co-purchase |
Without Pre-training |
60.5 |
59.9 |
58.7 |
90.13 |
Attribute masking |
63.2 |
58.8 |
62.2 |
90.98 |
Context Prediction |
58.6 |
64.3 |
/ |
/ |
For pre-training, we utilize the same pre-trained models from the paper, which have already pretrained on the large-scale ZINC15 and ChEMBL databases using a combination of node-level self-supervised strategies and graph-level supervised pre-training.
4.1.2 Datasets for extended evaluation
For the second part of our project, we evaluate the generalizability of the node-level pre-training strategies proposed in the paper on broader domains beyond chemistry and biology. We conduct experiments on different domains and several widely used datasets:
Cora, CiteSeer and PubMed the three datasets are citation network datasets, where nodes represent scientific publications, and edges represent citations between them. Each node has a feature vector representing the content of the publication and belongs to one of several classes representing research areas. We pretrain on other citation network datasets, Cora [10] and PubMed[11], and transfer the learned representations to CiteSeer[12]. PubMed has a larger number of nodes and classes. We combine Cora and PubMed for pre-training and transfer the learned representations to CiteSeer to evaluate the impact of using multiple source datasets.
Amazon Computers and Amazon Photo These two datasets are derived from the Amazon copurchase graph [13], where nodes represent products, and edges indicate that two products are frequently bought together. We pretrain on the Amazon Computers dataset and transfer the learned representations to the Amazon Photo dataset to assess the transferability of representations across different product domains.
4.2 Results
4.2.1 Reproducing
As shown in Table 1, the results from our replication study demonstrate the effectiveness of the pre-training strategies proposed in the paper. By comparing the performance of models with and without pre-training, we observe consistent improvements across all three datasets. This overall improvement highlights the potential of pre-training in enhancing the model’s performance, especially when the downstream dataset is relatively small, as is the case with ClinTox. Comparing the different pre-training strategies, we observe that the supervised pre-training approaches generally outperform their unsupervised counterparts. This suggests that incorporating task-specific supervision during pre-training can further improve the learned representations.
Overall, our replication study validates the findings of the original paper and demonstrates the effectiveness of the proposed pre-training strategies in improving the performance of GNN on molecular property prediction tasks. The consistent improvements across all three datasets highlight the potential of pre-training in enhancing the generalization ability and transferability of GNN in the chemistry domain.
4.2.2 Extended evaluation
From Table 2, the results of our experiments on broader domains demonstrate the effectiveness of the node-level pre-training strategies in improving the performance of GNN. For citation network datasets, Cora, CiteSeer, and PubMed, we observe that pre-training on Cora and transferring the learned representations to CiteSeer leads to improved performance on GCN and GIN compared to training on CiteSeer without pre-training. When we pretrain on the combination of Cora and PubMed datasets and transfer to CiteSeer, we see a further improvement in performance, indicating the benefits of using multiple source and larger datasets during pre-training.
In Amazon co-purchase datasets, Amazon computers and photo, pre-training on the Amazon computers dataset and transferring the learned representations to the Amazon Photo dataset also results ina significant performance boost compared to training on Amazon Photo without pre-training. This demonstrates the effectiveness of the pre-training strategies in capturing transferable knowledge across different product domains.
In brief, our experiments on broader datasets have shown the generalizability and effectiveness of the node-level pre-training strategies in improving the performance of GNN across various domains and tasks. The results highlight the potential of pre-training in enhancing the transferability of learned representations, which can reduce the reliance on large amounts of labeled data in the target domain.
5 Conclusion
In this project, we conducted a comprehensive study to evaluate the effectiveness and generalizability of the pre-training strategies for graph neural networks proposed in the paper. Our work consisted of two main parts: a replication study on molecular property prediction tasks to validate the findings of the original paper, and an evaluation of the pre-training strategies on broader domains beyond chemistry and biology. In the replication study, our results demonstrated overall improvements in performance across three datasets when using pre-trained GNN compared to the model without pre-training. These results prove the effectiveness of the proposed pre-training strategies in the chemistry domain. To assess the generalizability of the pre-training strategies, we conducted experiments on diverse datasets from other domains, including citation networks and co-purchase graphs. We evaluated the performance of GNN with and without pre-training and two node-level pre-training strategies, including Attribute Masking and Context Prediction. Our results showed that pre-training consistently improved the performance of GNN across the datasets in different domains, demonstrating the transferability of learned representations. Also, by combining multiple source datasets during pre-training, GNN probably can get a better performance.
In conclusion, our project provides strong evidence for the effectiveness and generalizability of the pre-training strategies proposed in the paper, which suggests that the pre-training strategy of GNN is a valuable tool for improving the performance in various domains.
References
[1]. Joan Bruna, Wojciech Zaremba, Arthur Szlam, and Yann LeCun. 2014. Spectral Networks and Locally Connected Networks on Graphs. In ICLR.
[2]. Weihua Hu, Bowen Liu, Joseph Gomes, Marinka Zitnik, Percy Liang, Vijay Pande, and Jure Leskovec. 2020. Strategies for Pre-Training Graph Neural Networks. In ICLR.
[3]. Yunshu Du, Wojciech M Czarnecki, Siddhant M Jayakumar, Mehrdad Farajtabar, Razvan Pascanu, and Balaji Lakshminarayanan. 2018. Adapting auxiliary losses using gradient similarity.1812.02224. In arXiv.
[4]. Zhao Chen, Vijay Badrinarayanan, Chen-Yu Lee, and Andrew Rabinovich. 2018. Gradnorm: Gradient normalization for adaptive loss balancing in deep multitask networks. 794–803. In ICML.
[5]. Alexandra Chronopoulou, Christos Baziotis, and Alexandros Potamianos. 2019. An embarrassingly simple approach for transfer learning from pretrained language models. 2089–2095. In NAACL.
[6]. Yi Yang, Cui, Hejie., Yang, Carl. (2023). PTGB: Pre-Train Graph Neural Networks for Brain Network Analysis. In Proceedings of Machine Learning Research - Conference on Health, Inference, and Learning 2023, 1–19. In CHIL.
[7]. Jacob Devlin, Ming-Wei Chang, Kenton Lee, and Kristina Toutanova. 2019. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. 4171–4186.In NAACL.
[8]. Nishai Kooverjee, Steven James, and Terence van Zyl. 2022. Investigating Transfer Learning in Graph Neural Networks. In International Conference on Learning Representations. In ICLR.
[9]. Zhenqin Wu, Bharath Ramsundar, Evan N Feinberg, Joseph Gomes, Caleb Geniesse, Aneesh S Pappu, Karl Leswing, and Vijay Pande. MoleculeNet: a benchmark for molecular machine learning. Chemical science, 9(2):513–530, 2018.
[10]. McCallum, A.K., Nigam, K., Rennie, J. et al. Automating the Construction of Internet Portals with Machine Learning. Information Retrieval 3, 127–163 (2000).
[11]. Sen, P., Namata, G., Bilgic, M., Getoor, L., Galligher, B., Eliassi-Rad, T. (2008). Collective Classification in Network Data. AI Magazine, 29(3), 93.
[12]. C. Lee Giles, Kurt D. Bollacker, and Steve Lawrence. 1998. CiteSeer: an automatic citation indexing system. In Proceedings of the third ACM conference on Digital libraries (DL ’98). Association for Computing Machinery, New York, NY, USA, 89–98.
[13]. Jaewon Yang and Jure Leskovec. 2012. Defining and evaluating network communities based on groundtruth. In Proceedings of the ACM SIGKDD Workshop on Mining Data Semantics (MDS ’12). Association for Computing Machinery, New York, NY, USA, Article 3, 1–8.
Cite this article
Li,Y.;Ge,Q. (2024). Enhancing graph neural network performance through comprehensive transfer learning strategies. Advances in Engineering Innovation,12,1-6.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Journal:Advances in Engineering Innovation
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Joan Bruna, Wojciech Zaremba, Arthur Szlam, and Yann LeCun. 2014. Spectral Networks and Locally Connected Networks on Graphs. In ICLR.
[2]. Weihua Hu, Bowen Liu, Joseph Gomes, Marinka Zitnik, Percy Liang, Vijay Pande, and Jure Leskovec. 2020. Strategies for Pre-Training Graph Neural Networks. In ICLR.
[3]. Yunshu Du, Wojciech M Czarnecki, Siddhant M Jayakumar, Mehrdad Farajtabar, Razvan Pascanu, and Balaji Lakshminarayanan. 2018. Adapting auxiliary losses using gradient similarity.1812.02224. In arXiv.
[4]. Zhao Chen, Vijay Badrinarayanan, Chen-Yu Lee, and Andrew Rabinovich. 2018. Gradnorm: Gradient normalization for adaptive loss balancing in deep multitask networks. 794–803. In ICML.
[5]. Alexandra Chronopoulou, Christos Baziotis, and Alexandros Potamianos. 2019. An embarrassingly simple approach for transfer learning from pretrained language models. 2089–2095. In NAACL.
[6]. Yi Yang, Cui, Hejie., Yang, Carl. (2023). PTGB: Pre-Train Graph Neural Networks for Brain Network Analysis. In Proceedings of Machine Learning Research - Conference on Health, Inference, and Learning 2023, 1–19. In CHIL.
[7]. Jacob Devlin, Ming-Wei Chang, Kenton Lee, and Kristina Toutanova. 2019. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. 4171–4186.In NAACL.
[8]. Nishai Kooverjee, Steven James, and Terence van Zyl. 2022. Investigating Transfer Learning in Graph Neural Networks. In International Conference on Learning Representations. In ICLR.
[9]. Zhenqin Wu, Bharath Ramsundar, Evan N Feinberg, Joseph Gomes, Caleb Geniesse, Aneesh S Pappu, Karl Leswing, and Vijay Pande. MoleculeNet: a benchmark for molecular machine learning. Chemical science, 9(2):513–530, 2018.
[10]. McCallum, A.K., Nigam, K., Rennie, J. et al. Automating the Construction of Internet Portals with Machine Learning. Information Retrieval 3, 127–163 (2000).
[11]. Sen, P., Namata, G., Bilgic, M., Getoor, L., Galligher, B., Eliassi-Rad, T. (2008). Collective Classification in Network Data. AI Magazine, 29(3), 93.
[12]. C. Lee Giles, Kurt D. Bollacker, and Steve Lawrence. 1998. CiteSeer: an automatic citation indexing system. In Proceedings of the third ACM conference on Digital libraries (DL ’98). Association for Computing Machinery, New York, NY, USA, 89–98.
[13]. Jaewon Yang and Jure Leskovec. 2012. Defining and evaluating network communities based on groundtruth. In Proceedings of the ACM SIGKDD Workshop on Mining Data Semantics (MDS ’12). Association for Computing Machinery, New York, NY, USA, Article 3, 1–8.









