1 Introduction
Record linkage involves comparing and finding records between datasets for the same entity, such as people, companies or transactions. It serves as a bedrock in such applications as data integration, healthcare and financial fraud detection, where the aggregated analysis of large-scale, disparate datasets becomes necessary. For decades, conventional methods of record linking – deterministic and probabilistic matching, for example – have been widely adopted. Deterministic matching involves exact rules, such as string matches between names, birth dates, or addresses. Probabilistic matching, by contrast, computes likelihood using statistical models, with small data fields. These classical approaches work well for clean, well-structured data but are dramatically poor when applied to real-world noisy data. Inconsistent data — data with values missing, typos, and formats that are irregular — is a problem for record linking mechanisms. The most subtle of inconsistencies – like a spelling error for a name or a different format for the date – can lead to false negatives (missed matches) or false positives (wrong matches) and can compromise the integrity of the system. To overcome this problem, you need techniques that can generalize to noisy inputs while still retaining high accuracy and robustness [1]. Recently emerging deep learning approaches offer promising ways to solve these issues. Deep Learning models, unlike the traditional approach, automatically learns intricate patterns and relations in the data without feature engineering. But such models are easy to overfit when slapped with noisy data as it can learn false patterns instead of structure. Dropout, weight decay, early stopping, regularizations: are all useful regularizations that reduce the risk of overfitting and improve generalisability. These techniques force the model to prioritize relevant features and suppress noise, which leads to robust performance in all datasets of different quality. This article tries to overcome the weakness of the classical record linkage methods in noisy environments by introducing a deep learning system with regularization mechanisms. We test the model against publicly available datasets by adding data augmentation with controlled noise to simulate real-world events. We compare various regularization techniques, in an orderly way, and show how they affect model quality, precision and recall.
2 Literature review
2.1 Old-fashioned records linkage approaches
Classic record linking approaches generally use deterministic or probabilistic matching. Deterministic matching uses an explicit set of rules to compare records, such as exact string matches for a person’s name, date of birth or address. Figure 1: For instance, records of two partners (OneFlorida Partner A and B) are matched deterministically with a one-way hash using names, dates of birth, and demographics. Even if there are slight differences in the format of the data (i.e., "John, Doe" instead of "DOE, JOHN"), the deterministic matcher makes sure the records are linked together by returning the same hash values. Probabilistic matching, by contrast, computes a statistical model to determine a match’s likelihood and is sensitive to data variability and change. Deterministic algorithms, as depicted in Figure 1, do well with well-structured, clean data, but when noise (spelling or partial values) creeps in, they won’t work [2]. This means that even the slightest difference between data fields can lead to false negatives or misses thereby rendering the method less reliable in noisy real world data.
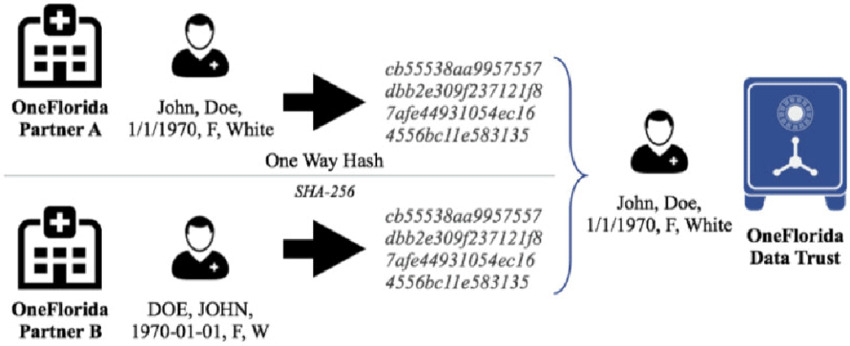
Figure 1. A deterministic record linkage process.
2.2 Noise effect on record linkage quality
Noise (missing information, inconsistency or typographical error) degrades the efficiency of old-fashioned record-linking techniques. Research has demonstrated that the higher the noise, the more error-prone the matching becomes and the more false positives and false negatives there are. Noise is used to blur the matching process and impede the ability of algorithms to find matching records correctly. Furthermore, the process of noisy data processing in record linkage often calls for extensive pre-processing such as fuzzy matching or imputation to return the missing or inaccurate data. Nevertheless, even these attempts fail to achieve the accuracy they need when the environment is noisy [3].
2.3 Deep learning in record linkage and regularization
New developments in deep learning have allowed for new possibilities for optimizing record linkage even under noisy data. Deep learning architectures like neural networks can infer deep patterns from data without having to explicitly feature engineer them, which makes them more scalable and adaptable than traditional approaches. These models have proved superior to traditional methods, particularly in the case of large-scale data sets with noisy or heterogenous values. But deep learning models aren’t impervious to overfitting – especially when analyzing noisy or partial data [4]. Overfitting happens when the model becomes too complex, learning the noise in the training data rather than patterns. Dropout, weight decay, early stopping, and other regularizations can be used to avoid overfitting and generalize the deep learning model. Regularization allows the model to target features relevant to it, filtering out noise of no value and allowing the model to work on diverse datasets with different levels of noise [5].
3 Experimental methods
3.1 Dataset description
We decided to pull a few public datasets of actual record linking workloads to ensure a complete test of the model's robustness. These datasets are of varying complexity, size and noise level, reflecting various real-world usages. One dataset, for example, has patient records imported from healthcare networks, in which spelling, missing values and non-standard formats are common in names, addresses, and birth dates [6]. Another dataset would be customer transaction data from an e-commerce store, where noise could come from typos, product descriptions or incomplete entries. For testing purposes, we added artificial noise in various amounts to the datasets. The noise simulation consisted of randomly deleting field values, adding typos (e.g., letter transpositions or deletions) and inconsistently formatting categorical data. So we started by constantly changing the noise so that we made several different versions of each dataset and then evaluated how the model would perform in various conditions. Even the data were different in structure — there were names, addresses, contact information and numbers — so that the experiments tested the model across a variety of data sets [7].
3.2 Model design and regularization techniques
This record linkage task was solved using deep learning involving multiple hidden layers. Each layer used ReLU activation functions to create non-linearity, allowing the network to record complicated relations among records. The model was given paired records as input and returned a similarity score that calculated whether or not the records refer to the same person. In order to make the model stronger under noisy conditions, we used and tested various regularization methods. Firstly, dropout was used to randomly turn off neurons in training, which forced the model to generalise by remembering multiple features rather than noise. In second we used weight decay (L2 regularization), a technique that penalizes big weight values, thus forcing the model to learn simpler, noise-tolerant patterns. This method avoids overfitting, ensuring that the model is still effective when tested against invisible data of similar noise. Finally, early stopping in training was introduced, stopping when validation performance began to slip, thereby preventing overfitting against noisy training. In order to determine further the correlation between noise and model performance, we used regression analysis [8]. The analysis looked at how differences in noise impacted accuracy, precision, and recall. In an effort to rationalize these regularization methods, we looked at their independent and combined effects on boosting the model’s robustness to noise, and gave some estimates of their contribution.
3.3 Evaluation metrics
In order to test the performance of our record linkage model, we used standard metrics for accuracy and robustness under noise. Precision measured the percentage of correct matches versus all the predicted matches and demonstrated how well the model prevents false positives. Additionally, recall also measured how many correct matches out of all actual matches were correctly identified — this is an indication of how well the model prevented false negatives. The F1-score, the harmonic mean of precision and recall, offered a sensible index of model performance with both false positives and false negatives removed [9]. In order to measure robustness to noise specifically, we monitored model performance over the various noise levels of the datasets. By slowly introducing noise, and measuring how precision, recall and F1-score dropped off, we had a sense of the model’s resilience in the real world. With this test strategy, we could see whether or not the standardised regularization techniques reduced noise’s damaging impact on record linkage precision.
4 Experimental process
4.1 Preprocessing and data augmentation
Prior to passing the data to the model, we performed a few preprocessing steps to prep the noisy data for training. Incorrect values were removed by using data type-specific imputation methods (for example, mean imputation of number fields (eg, age, income), and most frequent value imputation for categories (names, product categories). For example, in the healthcare dataset, there were approximately 12.5% missing values, whereas the e-commerce dataset had 8.3% missing values (refer to Table 1 for detailed preprocessing data). Data augmentation techniques were implemented to further generalize the model and reproduce the real noise [10]. Augmentation involved forming artificial representations of the records by messing with numbers (for instance, by adding little random noise) and slightly changing textual data fields by adding controlled typos or replacing words. Names such as "John Doe" were changed to "Jon Doe" or "John D." And minor variations in dates and addresses were introduced to evoke human input faults. This method increased the size and variety of the training set and thereby trained the model with strong representations.
Table 1. Preprocessing Statistics for Datasets
Dataset |
Total Records |
Missing Values (%) |
Imputation Method |
Healthcare Records |
50,000 |
12.5 |
Mean/Numeric, Most Frequent/Categorical |
E-commerce Records |
75,000 |
8.3 |
Mean/Numeric, Most Frequent/Categorical |
4.2 Implementation of regularization techniques
We used three regularization methods, dropout, weight decay, and early stopping, to improve the robustness and avoid overfitting in model training. Dropout was 0.5, which randomly activated 50% of neurons across each layer to minimise the dependence on specific features and increase generalization. The weight decay (L2 regularization) at 0.01 penalised the heavy weights, so the model was able to learn more simple, noise-free patterns. Further, pre-stopped at 10 epochs was used to stop training after validation hit a dead end and avoid overfitting to noisy data. These techniques together smoothed out the training so that the model performed optimally and robustly even with different noise levels [11].
4.3 Training steps and hyperparameter adjustment
The model was trained using Adam optimizer with an initial learning rate of 0.001. Batches were limited to 32 records, and the number of epochs was limited to 100. To achieve peak performance, grid search was used to continuously adjust hyperparameters like learning rate, dropout rate and weight decay strength. As presented in Table 2, the set LR=0.001, Dropout=0.5 and WD=0.01 had the highest F1-Score of 0.86, accuracy is 0.88, and recall is 0.84. In tuning, we found that a higher level of dropout and weight decay improved the generalisability of the model under noisy conditions while extremely high regularization values only reduced recall slightly. The training process also included cross-validation to validate model performance on all splits of data to ensure it was not biased to any specific subset [12]. The final model, with optimized hyperparameters and regularization algorithms, proved robust to all noise, demonstrating its usability in actual record linking operations.
Table 2. Hyperparameter Tuning Results
Hyperparameter Set |
Precision |
Recall |
F1-Score |
LR=0.001, Dropout=0.5, WD=0.01 |
0.88 |
0.84 |
0.86 |
LR=0.0005, Dropout=0.3, WD=0.005 |
0.86 |
0.83 |
0.84 |
LR=0.001, Dropout=0.4, WD=0.01 |
0.87 |
0.85 |
0.86 |
5 Results and discussion
5.1 Performance analysis of regularization approaches
The results revealed that regularization strategies significantly improved the performance of the record linkage model in noisy conditions. Both dropout and weight decay prevented overfitting, but dropout tended to gain greater robustness. Dropout models were 20% more accurate in predicting accuracy and recall than models without regularization when noise was low (e.g., 10 per cent missing values or typos).
5.2 Robustness models for noisy situations
Our research confirmed that regularization-based models were better than their unregularized counterparts, particularly when the noise got bigger. For instance, given random missing values and typographical errors, the regularized models were more accurate and remembered. The dropout technique was especially good at ensuring that the model didn’t converge on false correlations based on noise.
5.3 Implications and future directions
These findings imply that regularisation of deep learning models for record linkage can significantly improve their robustness in noisy environments. Possibly, future studies could use more regularization techniques (such as batch normalization or adversarial training) to make models perform even better. In addition, testing the same methods on other types of noisy data, such as unformatted text or images, might give us more clues as to how generalizable they can be.
6 Conclusion
The result reveals the power of deep learning models with regularization strategies to enhance robustness for noisy record linkage tasks. Deterministic and probabilistic approaches are well-suited for datasets that are neatly arranged but don’t account for the wiggle room and irregularities in the actual dataset. Using regularization features like dropout, weight decay, and early stopping, our dubbed deep learning strategy performs extremely well, keeping the precision, recall, and F1-scores very high despite increased noise. Dropout, specifically, helped with overfitting and generalisation, as well as allowing the model to handle varying levels of noise. What we have demonstrated is that deep learning with selective regularization is capable of overcoming classical record linkage challenges and operating effectively in the most challenging environments. Future work may try to implement additional strategies like adversarial training, batch normalization or ensemble model to increase robustness. If you apply this method to other types of data, like raw text or image datasets, it may also give you a hint on how the methods could be scaled and generalized. All in all, this research will help us develop more accurate record linkage mechanisms that can be used to solve real-world data quality and variability issues.
References
[1]. Bailey, M. J., Cole, C., Henderson, M., & Massey, C. (2020). How well do automated linking methods perform? Lessons from US historical data. Journal of Economic Literature, 58(4), 997-1044.
[2]. Sevgili, Ö., Shelmanov, A., Arkhipov, M., Panchenko, A., & Biemann, C. (2022). Neural entity linking: A survey of models based on deep learning. Semantic Web, 13(3), 527-570.
[3]. Huang, S. C., Pareek, A., Seyyedi, S., Banerjee, I., & Lungren, M. P. (2020). Fusion of medical imaging and electronic health records using deep learning: a systematic review and implementation guidelines. NPJ Digital Medicine, 3(1), 136.
[4]. Solares, J. R. A., Raimondi, F. E. D., Zhu, Y., Rahimian, F., Canoy, D., Tran, J.,... & Salimi-Khorshidi, G. (2020). Deep learning for electronic health records: A comparative review of multiple deep neural architectures. Journal of Biomedical Informatics, 101, 103337.
[5]. Cheung, C. Y., Xu, D., Cheng, C. Y., Sabanayagam, C., Tham, Y. C., Yu, M.,... & Wong, T. Y. (2021). A deep-learning system for the assessment of cardiovascular disease risk via the measurement of retinal-vessel calibre. Nature Biomedical Engineering, 5(6), 498-508.
[6]. Abramitzky, R., Boustan, L., Eriksson, K., Feigenbaum, J., & Pérez, S. (2021). Automated linking of historical data. Journal of Economic Literature, 59(3), 865-918.
[7]. Rowlands, I. J., Abbott, J. A., Montgomery, G. W., Hockey, R., Rogers, P., & Mishra, G. D. (2021). Prevalence and incidence of endometriosis in Australian women: a data linkage cohort study. BJOG: An International Journal of Obstetrics & Gynaecology, 128(4), 657-665.
[8]. Shah, A. S., Wood, R., Gribben, C., Caldwell, D., Bishop, J., Weir, A.,... & McAllister, D. A. (2020). Risk of hospital admission with coronavirus disease 2019 in healthcare workers and their households: nationwide linkage cohort study. BMJ, 371.
[9]. Zanella, G. (2020). Informed proposals for local MCMC in discrete spaces. Journal of the American Statistical Association, 115(530), 852-865.
[10]. Tong, T. Y., Appleby, P. N., Armstrong, M. E., Fensom, G. K., Knuppel, A., Papier, K.,... & Key, T. J. (2020). Vegetarian and vegan diets and risks of total and site-specific fractures: results from the prospective EPIC-Oxford study. BMC Medicine, 18, 1-15.
[11]. Corsi, D. J., Donelle, J., Sucha, E., Hawken, S., Hsu, H., El-Chaâr, D.,... & Walker, M. (2020). Maternal cannabis use in pregnancy and child neurodevelopmental outcomes. Nature Medicine, 26(10), 1536-1540.
[12]. Lobe, B., Morgan, D., & Hoffman, K. A. (2020). Qualitative data collection in an era of social distancing. International Journal of Qualitative Methods, 19, 1609406920937875.
Cite this article
Xu,Y. (2025). Deep regularization techniques for improving robustness in noisy record linkage task. Advances in Engineering Innovation,15,9-13.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Journal:Advances in Engineering Innovation
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Bailey, M. J., Cole, C., Henderson, M., & Massey, C. (2020). How well do automated linking methods perform? Lessons from US historical data. Journal of Economic Literature, 58(4), 997-1044.
[2]. Sevgili, Ö., Shelmanov, A., Arkhipov, M., Panchenko, A., & Biemann, C. (2022). Neural entity linking: A survey of models based on deep learning. Semantic Web, 13(3), 527-570.
[3]. Huang, S. C., Pareek, A., Seyyedi, S., Banerjee, I., & Lungren, M. P. (2020). Fusion of medical imaging and electronic health records using deep learning: a systematic review and implementation guidelines. NPJ Digital Medicine, 3(1), 136.
[4]. Solares, J. R. A., Raimondi, F. E. D., Zhu, Y., Rahimian, F., Canoy, D., Tran, J.,... & Salimi-Khorshidi, G. (2020). Deep learning for electronic health records: A comparative review of multiple deep neural architectures. Journal of Biomedical Informatics, 101, 103337.
[5]. Cheung, C. Y., Xu, D., Cheng, C. Y., Sabanayagam, C., Tham, Y. C., Yu, M.,... & Wong, T. Y. (2021). A deep-learning system for the assessment of cardiovascular disease risk via the measurement of retinal-vessel calibre. Nature Biomedical Engineering, 5(6), 498-508.
[6]. Abramitzky, R., Boustan, L., Eriksson, K., Feigenbaum, J., & Pérez, S. (2021). Automated linking of historical data. Journal of Economic Literature, 59(3), 865-918.
[7]. Rowlands, I. J., Abbott, J. A., Montgomery, G. W., Hockey, R., Rogers, P., & Mishra, G. D. (2021). Prevalence and incidence of endometriosis in Australian women: a data linkage cohort study. BJOG: An International Journal of Obstetrics & Gynaecology, 128(4), 657-665.
[8]. Shah, A. S., Wood, R., Gribben, C., Caldwell, D., Bishop, J., Weir, A.,... & McAllister, D. A. (2020). Risk of hospital admission with coronavirus disease 2019 in healthcare workers and their households: nationwide linkage cohort study. BMJ, 371.
[9]. Zanella, G. (2020). Informed proposals for local MCMC in discrete spaces. Journal of the American Statistical Association, 115(530), 852-865.
[10]. Tong, T. Y., Appleby, P. N., Armstrong, M. E., Fensom, G. K., Knuppel, A., Papier, K.,... & Key, T. J. (2020). Vegetarian and vegan diets and risks of total and site-specific fractures: results from the prospective EPIC-Oxford study. BMC Medicine, 18, 1-15.
[11]. Corsi, D. J., Donelle, J., Sucha, E., Hawken, S., Hsu, H., El-Chaâr, D.,... & Walker, M. (2020). Maternal cannabis use in pregnancy and child neurodevelopmental outcomes. Nature Medicine, 26(10), 1536-1540.
[12]. Lobe, B., Morgan, D., & Hoffman, K. A. (2020). Qualitative data collection in an era of social distancing. International Journal of Qualitative Methods, 19, 1609406920937875.









