1. Introduction
In recent times, medical imaging's transformative potential has been at the forefront of global health discussions, with lung cancer diagnosis being of paramount significance. Lung cancer, notoriously known for its high mortality rates, underscores the urgency of developing advanced diagnostic tools [1-3]. Fortunately, recent progress in artificial intelligence, particularly in the field of deep learning, holds immense potential for revolutionizing this field. Its potential to enhance the intricacy and scope of medical imaging is truly remarkable.
Generative modeling, a subset of deep learning, provides powerful tools for creating synthetic yet realistic images [4-7]. Among the vast array of generative models, two have emerged as frontrunners in terms of their applicability and promise, namely the Variational Autoencoder (VAE) and the Diffusion model.
Irem Cetin, Maialen Stephens et al proposed an improved deep learning model called Attri-VAE to better understand and explain the relationship between medical images and clinical data. When this method was applied to analyze medical data of healthy people and patients with myocardial infarction, it not only reconstructed with high accuracy, but also distinguished and interpreted different data characteristics more clearly, surpassing the existing technology [8].
In terms of diffusion models, Firas Khader, Gustav Müller-Franzes and others explored the potential of diffusion models to generate 3D data in the medical field. They propose a new diffusion model architecture specifically for the latent space of 3D medical images. They trained using fthepublicly available datasets and checked whether the generated images looked realistic and quantified their diversity. Finally, they show practical applications of using these synthetic images in clinical settings, especially in data-limited settings, and whether image segmentation models can be improved by pre-training synthetic images [9].
However, while both models have showcased individual prowess in various domains, a comprehensive and side-by-side comparison in the context of lung cancer imaging has not been extensively investigated. Such a comparative exploration is not merely academic. It has profound implications for clinical applications. By understanding the strengths, weaknesses, and nuances of each model, medical practitioners and researchers can tailor their approaches more effectively, optimizing the diagnostic process and potentially saving countless lives.
This paper embarks on this exploratory journey, providing a deep dive into the capabilities of VAEs and Diffusion models as they are applied to lung cancer imagery.
2. Method
2.1. Dataset preparation
The data source is the "The Iraq-Oncology Teaching Hospital/National Center for Cancer Diseases (IQ-OTH/NCCD) lung cancer dataset." This dataset was collected from the Iraq-Oncology Teaching Hospital and the National Center for Cancer Diseases over a three-month period in fall 2019. The dataset includes CT scans of individuals diagnosed with lung cancer at various stages, as well as healthy subjects. The CT scans were marked by oncologists and radiologists from these two centers. The dataset consists of a total of 1190 images, representing CT scan slices from 110 cases [10].
Figure 1. The sample images used in this study.
2.2. Model
2.2.1. VAE
Introduction of VAE. A Variational Autoencoder (VAE) is a generative model that, like traditional autoencoders, can encode and decode data. However, its unique feature is that it learns a probabilistic representation of the data, making it capable of generating new, similar samples.
Basic Structure of a VAE. Encoder: Takes an input sample and transforms it into two things— a mean and a variance. These together define a probabilistic distribution in the latent space.
Latent Space: A continuous space where the probabilistic distribution defined by the encoder's outputs exists. Decoder: Samples a point from the latent space and decodes it into an output sample.
The structure of the VAE (The following is the complete process). In the study,a Variational Autoencoder (VAE) was used to model and generate medical imaging data, following a well-structured pipeline. Here's an overview of the code structure:
Data Import and Decompression. This study initiated by storing the dataset on Google Drive, a cloud storage solution, for efficient handling in a Google Colab environment. This choice offers a seamless way to import data, eliminating dependencies on local storage. The dataset is housed as a zip file, and with clearly defined path parameters, it is directly decompressed from the cloud drive, paving the way for subsequent image loading and processing.
Image Preprocessing & Loading. Given the potential variations in size and formats of medical images, a preprocessing phase is introduced to ensure input consistency. The load_image function is tasked with converting images to the RGB format and standardizing their sizes, ensuring all images are uniformly treated by the model. By iterating through the data directory, the load_data function successfully loads the entire dataset into a list of processed images.
Data Normalization. For the stability and efficacy of model training, all image data are transformed into NumPy array format and normalized to range between 0 and 1. This normalization strategy offers numerically stable data for model training, mitigating issues like gradient vanishing or explosion.
Model Architecture Definition. The VAE model architecture encompasses the encoder, decoder, and the overarching VAE structure:
Encoder. Employing a series of reducing convolution operations, the encoder maps image data to a latent space, producing vectors that represent the image content. Additionally, the encoder outputs mean and log variance, both of which play a crucial role in subsequent random sampling.
Decoder. Accepts latent vectors produced by the encoder and reconstructs the original image data through deconvolution operations. This step forms the basis for new image generation.
Overall Architecture. Integrating the encoder and decoder, it defines the flow from input images to latent vectors and back to regenerated images. This structure also touches upon the loss computation, which includes reconstruction loss and KL divergence.
Model Training. This study opted for the Adam optimizer for model training and set a training duration of 200 epochs. These settings were derived from prior experiments and best practices, aiming to offer optimal model parameters for medical image generation.
New Image Generation. Once the VAE model is trained, it can be leveraged to generate new medical images. This study introduced a random sampling strategy to draw latent vectors from a standard normal distribution and use the decoder for image generation. These images offer a visual assessment of the model's generative capabilities.
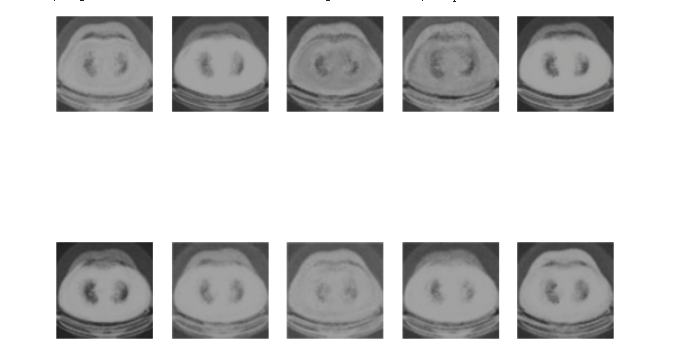
Figure 2. The generated image by VAE.
Through this structure, this study provided a systematic approach to employing VAEs for medical image generation, laying a robust foundation for future research and applications. Figure 2 shows the medical image of lung cancer generated by the VAEs model.
2.2.2. Diffusion model
Introduction of Diffusion model
The diffusion model, an emerging paradigm in the realm of deep learning, encapsulates a method of iterative data transformation by emulating a process akin to diffusion. Rooted in the physical world, diffusion pertains to the manner in which substances spread through space and time, predominantly due to random motion of molecules. Adapting this concept to the digital domain, the diffusion model leverages this phenomenon to generate or reconstruct data iteratively.
In applications like image generation, instead of producing an image outright, the model starts with a noisy version and gradually refines it, leading to the final desired outcome. This nuanced procedure contrasts with many traditional generative models that might produce outputs in one go.
The structure of the Diffusion model
Data Import and Decompression. To ensure efficient data handling, this study operated within the Google Colab environment, storing the requisite medical imagery dataset on Google Drive. Via a designated path, the dataset is imported as a zip file and directly decompressed from the cloud, ensuring seamless subsequent handling.
Image Preprocessing and Loading. Medical imagery often varies in dimensions and formats. To maintain data uniformity, we've incorporated a preprocessing step. The load_image function ensures every image transitions to an RGB format and conforms to a specified size. Subsequently, the load_data function iterates through the defined data directory, loading and generating a list of preprocessed images.
Model Structural Definition. The controlled diffusion model is designed to iteratively remove image noise while progressively restoring the original image details:
Diffusion Mechanism. Based on a mathematical model, this mechanism simulates the potential multilayered noise effects on images by iteratively introducing Gaussian noise. Each noise iteration builds upon the preceding result, ensuring layered noise introduction.
Model Core. At the heart of the model is a U-Net-like structure, particularly apt for image segmentation and restoration tasks. It comprises an encoding and decoding pathway, capturing and reconstructing image characteristics and minutiae.
Loss Function. Training the model focuses on minimizing the divergence between the reconstructed and original images. This study employed the Mean Squared Error (MSE) to gauge this discrepancy and optimize the model weights.
Model Training. Given the intricacy and noise distribution of medical imagery, we've chosen the Adam optimizer, setting a distinct training epoch duration. Moreover, this study validated model performance post each epoch, ensuring not just noise removal but also the retention of primary image features and details.
Image Restoration and Generation. Upon concluding training, the controlled diffusion model can be used to restore damaged medical images. This study introduced noisy images to the model, anticipating clear, noise-free medical image outputs.
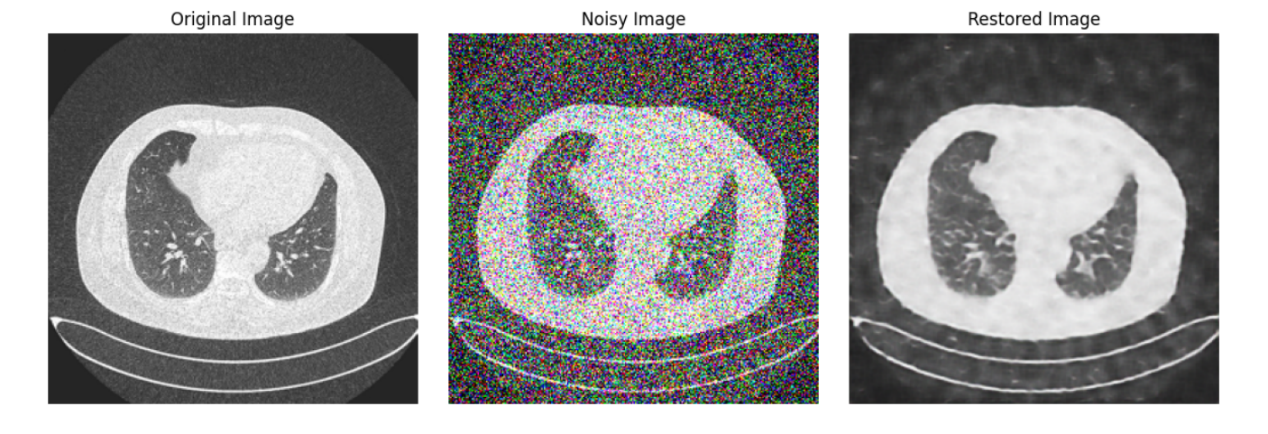
Figure 3. The medical images generated by the diffusion model.
Through the structured delineation above, we've vividly showcased how to leverage the controlled diffusion model technique for medical image data handling, aimed at enhancing image quality and eliminating noise. This lays a robust foundation for subsequent research and applications in medical imagery. Figure 3 shows the medical image of lung cancer generated by the Diffusion model.
2.3. Implementation details
2.3.1. VAE Model
Architecture
Encoder: Takes an input image and encodes it into two vectors: a mean vector (μ) and a standard deviation vector (σ). Typically, convolutional layers are used for image data. Decoder: Takes a latent vector sampled from the distribution defined by (μ, σ) and decodes it to generate an image. Deconvolutional (or upsampling) layers are usually employed for this task.
Training
Loss function: Combines a reconstruction term (like Mean Squared Error between the input and its reconstruction) and a regularization term (KL-divergence between the encoded distribution and a standard normal distribution).
Backpropagation & Optimization: The loss is minimized using an optimizer like Adam.
Sampling
To generate new samples, sample a vector from a standard normal distribution and pass it through the decoder.
2.3.2. Diffusion model
Model Dynamics
The model works by starting with a target data point and progressively adding noise in reverse to arrive at a noised version. During generation, the model does the opposite, removing noise step by step until the desired data is constructed.
Architecture
Typically involves a neural network that predicts the parameters of the noise process. This network conditions its predictions on both the current data state and the step number in the diffusion process.
Training
Loss function: Usually Mean Squared Error between the true data and the data generated by following the diffusion process backwards.
Backpropagation & Optimization: Gradient-based optimization techniques are used to update the model parameters.
Sampling
Generation is done by starting with a random noise sample and running the diffusion process forwards, removing noise at each step.
Commonalities
Both VAE and Diffusion Model are unsupervised learning models used for generative tasks. They both depend heavily on stochastic processes: VAE on sampling from the latent space and the Diffusion Model on the noise addition/removal process. Both models benefit from using deep neural networks as their backbone.
Differences
While VAEs have a clear encoder-decoder architecture, diffusion models use the same model structure for both corruption (adding noise) and denoising (removing noise).
The training dynamics and objective functions are different. VAEs aim to match the latent distribution to a prior (often Gaussian), while diffusion models aim to learn the parameters of a complex noise process.
Implementing these models requires a strong foundation in deep learning, proficiency with deep learning frameworks like TensorFlow or PyTorch, and an understanding of the respective model's dynamics.
Using 200 Epoch to run the VAE model, and I only use 10 Epoch to run the Diffusion model.
Image analysis of the two models
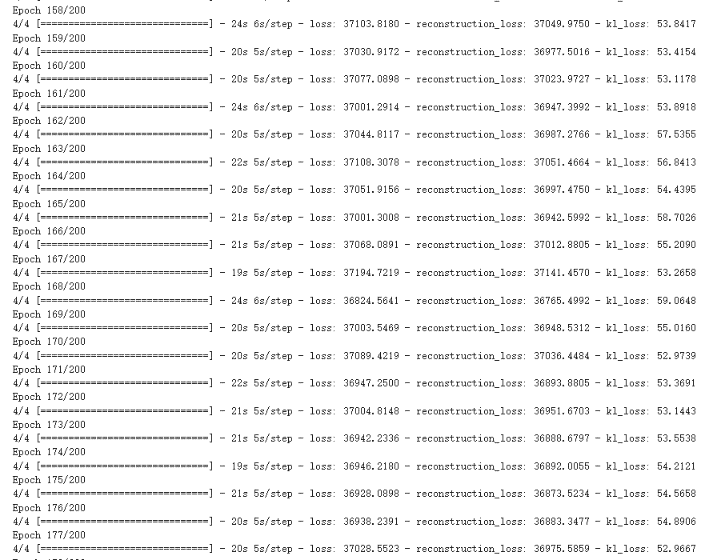
Figure 4. The training process of VAE.
For the lung cancer pictures generated by the VAE model, it can be found out by observing the picture Figure 2. The picture is more blurred than the original version. It can be observed that a lung cancer on the picture, but it basically can't see too many details. However, the VAE model generates images faster, and the average time to complete an Epoch is about 20 seconds (Figure 4), and it can be mass-produced quickly.
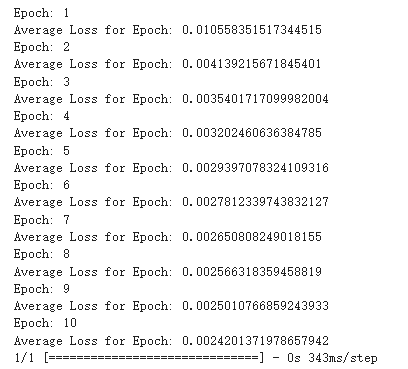
Figure 5. The training process of diffusion model.
By observing the image generated by the diffusion model, it can be observed that it has more and better details, and it only uses 10 Epoch for training shown in Figure 5. It almost perfectly imitates the original image, although it has a huge flaw, that is the generation speed is very slow.
3. Results and discussion
The primary objective was to provide a comprehensive comparison between the Variational Autoencoder (VAE) and the Diffusion model in the context of generating lung cancer images. Herein, this study highlighted the results and share insights based on the observations.
Results of Image Quality and Details. VAE: The images generated by the VAE, as seen in Figure 2, were comparatively blurrier and lacked the intricate details seen in the original images. However, the VAE was able to roughly capture the primary feature of lung cancer, presenting a decent approximation.
Diffusion Model: The diffusion model-generated images, depicted in Figure 3, showcased superior fidelity. These images retained a significant amount of the details seen in the original lung cancer CT scans, thereby demonstrating its ability to generate more clinically useful images.
Training and Generation Time. VAE: The VAE model's training efficiency was evident. With 200 epochs, the average time per epoch was roughly 20 seconds, as indicated in Figure 4. This brisk training, coupled with its faster image generation speed, implies that the VAE could be advantageous in scenarios demanding rapid image production.
Diffusion Model: While the diffusion model required a mere 10 epochs, its generative process was notably slower. Although this model delivers superior image quality, its prolonged generation time might be a bottleneck in certain applications.
Epochs and Convergence:
Discussion. Model Selection and Use Cases: The choice between VAE and the Diffusion model largely hinges on the specific demands of the application. For real-time or rapid diagnostics, where time is crucial, VAEs might be preferred, despite the compromise on image quality. On the other hand, in research settings or cases where high-fidelity images are paramount, the Diffusion model is the clear winner.
Potential Improvements: The VAE's performance might be enhanced by employing more advanced architectures or integrating regularization techniques. Meanwhile, the speed bottleneck of the Diffusion model might be addressed by parallelizing the generative process or employing more efficient hardware.
Future Prospects: Given the burgeoning advancements in deep learning, this study envision hybrid models that amalgamate the strengths of both VAEs and diffusion models. Such models could potentially offer both speed and image quality, thereby serving a wider array of medical imaging applications.
4. Conclusion
This research paper conducts a comparative examination of VAE model and Diffusion model, specifically in the context of lung cancer imaging. Utilizing the "Iraq-Oncology Teaching Hospital/National Center for Cancer Diseases (IQ-OTH/NCCD) lung cancer dataset" as the data source, the study scrutinizes the effectiveness, precision, and faithfulness of the images produced by each model. In conclusion, both VAE and the Diffusion model offer distinct advantages in the domain of lung cancer image generation. While VAE prioritizes speed, the Diffusion model champions image quality. Determining which model to employ is contingent upon the specific requirements of the task at hand. Through this study, further study hopes to guide medical practitioners and researchers in their endeavors, striving for early and accurate lung cancer diagnostics.
References
[1]. Minna J D Roth J A and Gazdar A F 2002 Focus on lung cancer. Cancer cell 1(1) 49-52
[2]. Schabath M B and Cote M L 2019 Cancer progress and priorities: lung cancer Cancer epidemiology, biomarkers & prevention 28(10) 1563-1579
[3]. Spiro S G and Silvestri G A 2005 One hundred years of lung cancer. American journal of respiratory and critical care medicine 172(5) 523-529
[4]. Creswell A et al 2018 Generative adversarial networks: An overview IEEE signal processing magazine 35(1) 53-65
[5]. Wang K et al 2017 Generative adversarial networks: introduction and outlook IEEE/CAA Journal of Automatica Sinica 4(4) 588-598
[6]. Goodfellow I et al 2020 Generative adversarial networks Communications of the ACM 63(11) 139-144
[7]. Aggarwal A et al 2021 Generative adversarial network: An overview of theory and applications. International Journal of Information Management Data Insights 1(1) 100004
[8]. Cetin I et al 2023 Attri-VAE: Attribute-based interpretable representations of medical images with variational autoencoders Computerized Medical Imaging and Graphics 104 102158
[9]. Khader F et al 2023 Denoising Diffusion Probabilistic Models for 3D Medical Image Generation Nature News Nature Publishing Group 5 May 2023
[10]. Al-Yasriy Hamdalla F 2020 The IQ-Oth/NCCD Lung Cancer Dataset Kaggle 24 May 2020 www.kaggle.com/datasets/hamdallak/the-iqothnccd-lung-cancer-dataset
Cite this article
Chen,Z. (2024). Comparison of VAE model and diffusion model in lung cancer images generation. Applied and Computational Engineering,47,28-35.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 4th International Conference on Signal Processing and Machine Learning
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Minna J D Roth J A and Gazdar A F 2002 Focus on lung cancer. Cancer cell 1(1) 49-52
[2]. Schabath M B and Cote M L 2019 Cancer progress and priorities: lung cancer Cancer epidemiology, biomarkers & prevention 28(10) 1563-1579
[3]. Spiro S G and Silvestri G A 2005 One hundred years of lung cancer. American journal of respiratory and critical care medicine 172(5) 523-529
[4]. Creswell A et al 2018 Generative adversarial networks: An overview IEEE signal processing magazine 35(1) 53-65
[5]. Wang K et al 2017 Generative adversarial networks: introduction and outlook IEEE/CAA Journal of Automatica Sinica 4(4) 588-598
[6]. Goodfellow I et al 2020 Generative adversarial networks Communications of the ACM 63(11) 139-144
[7]. Aggarwal A et al 2021 Generative adversarial network: An overview of theory and applications. International Journal of Information Management Data Insights 1(1) 100004
[8]. Cetin I et al 2023 Attri-VAE: Attribute-based interpretable representations of medical images with variational autoencoders Computerized Medical Imaging and Graphics 104 102158
[9]. Khader F et al 2023 Denoising Diffusion Probabilistic Models for 3D Medical Image Generation Nature News Nature Publishing Group 5 May 2023
[10]. Al-Yasriy Hamdalla F 2020 The IQ-Oth/NCCD Lung Cancer Dataset Kaggle 24 May 2020 www.kaggle.com/datasets/hamdallak/the-iqothnccd-lung-cancer-dataset









