1. Introduction
The human brain is a vital and complex organ, and brain tumors are not only prevalent but also have a high mortality rate. Some malignant brain tumors can develop into cancer. An estimated 87,500 new cancer cases (42,400 males and 45,100 females) occurred in 2022 [1]. For patients with brain tumors, accurately delineating the tumor's location in MRI scans is crucial for effective treatment planning. The conventional practice for segmenting tumor areas is manual segmentation. However, this method is time-consuming, labor-intensive, and the results varies among observers [2]. Consequently, this underscores the importance of integrating automated segmentation techniques into brain tumor MRI analysis, which could significantly enhance efficiency and accuracy in clinical practice.
Utilizing strong magnetic fields and radiofrequency waves, MRI is particularly effective in imaging soft tissues, including muscles, brains, and internal organs, which are not easily discernible with conventional radiographic methods like X-rays. The dataset utilized in this study, derived from the BraTS 2020, includes four distinct MRI modalities, each designed to emphasize different aspects of brain anatomy and pathology.
MRI, by utilizing strong magnetic fields and radiofrequency waves, excels at imaging soft tissues such as the brain, muscles, and internal organs—regions not easily captured by traditional radiographic methods like X-rays. The dataset utilized in this study, BraTS 2020, comprises four distinct MRI modalities, each highlighting different aspects of brain anatomy and pathology.
The MRI dataset consists of 4 modalities. Fluid-Attenuated Inversion Recovery (FLAIR) suppresses cerebrospinal fluid (CSF) signals, enhancing lesion visibility near CSF. T1-Weighted Imaging (T1) highlights fatty tissue, offering detailed anatomical brain assessment. When combined with a contrast agent, T1-Weighted Imaging with Contrast Enhancement (T1CE) further improves the visibility of specific tissues and lesions, enabling more precise diagnoses. T2-Weighted Imaging (T2), emphasizing fluid and edema, is essential for depicting tissue water content and contrasting soft tissues. The MRI data also contains a segmentation image, which serves as the ground truth for training and validation of deep learning models.
2. Literature review
Deep learning has driven significant advancements in medical image segmentation, particularly in structured tasks like brain tumor segmentation. While architectures such as U-Net have been effective in capturing local features and hierarchical representations, they face limitations in retaining fine spatial details, especially when segmenting small or irregular structures [3]. Conventional CNNs and Fully Convolutional Neural Networks (FCNNs) often lack the ability to capture multi-scale information effectively, leading to suboptimal performance when dealing with objects of varying sizes and complex textures [4,5,6]. These methods also struggle to incorporate global contextual information, critical for improving segmentation accuracy in challenging scenarios.
This study addresses these challenges by introducing an enhanced segmentation framework based on the DeepLabv3+ architecture, which utilizes atrous convolution and Atrous Spatial Pyramid Pooling (ASPP) to capture multi-scale context and retain fine spatial details, improving boundary precision and overall segmentation performance [7].
3. Methodology
The DeepLabv3+ architecture is specifically designed for high-resolution semantic segmentation, leveraging a combination of advanced feature extraction and precise upsampling techniques. This architecture is composed of a decoder and an encoder. The encoder in DeepLabv3+ is tasked with extracting features from the input image. Leveraging specialized atrous convolutions and the atrous spatial pyramid pooling (ASPP) structure, DeepLabv3+ captures multi-scale contextual information while preserving high-resolution features. The decoder then enhances the segmentation output by recovering spatial details lost during encoding, creating a detailed and accurate segmentation map that integrates high-level semantic information with fine-grained spatial details. By integrating atrous convolution with multi-scale feature extraction, DeepLabv3+ effectively captures detailed contextual information from the brain slices while maintaining spatial resolution.
3.1. Atrous convolution
The atrous convolution is the basic component of ASPP. Within this framework, the atrous convolution is employed to enhance the encoder's ability to capture features across multiple scales.
In contrast to standard convolution, atrous convolution incorporates a dilation rate, which broadens the receptive field without raising the number of parameters or sacrificing spatial resolution. The operation of atrous convolution can be represented by the following equation:
\( y[i]=\sum _{k}x[i+r∙k]w[k]\ \ \ (1) \)
where \( y[i] \) denotes the value at position i in the output, \( x[i+r∙k] \) represents the corresponding value in the input, and \( w[k] \) refers to the weight of the convolution kernel at index k.
The dilation rate r defines the stride for sampling the input data. In this model, dilation rates of 6, 12, and 18 were used to accommodate various sizes of brain tumors. Atrous convolution is identical to standard convolution when the dilation rate is set to 1. Compared to standard convolution, atrous convolution offers superior flexibility in segmenting brain tumors of various sizes due to its adaptable receptive field. An illustration is shown in Figure1.
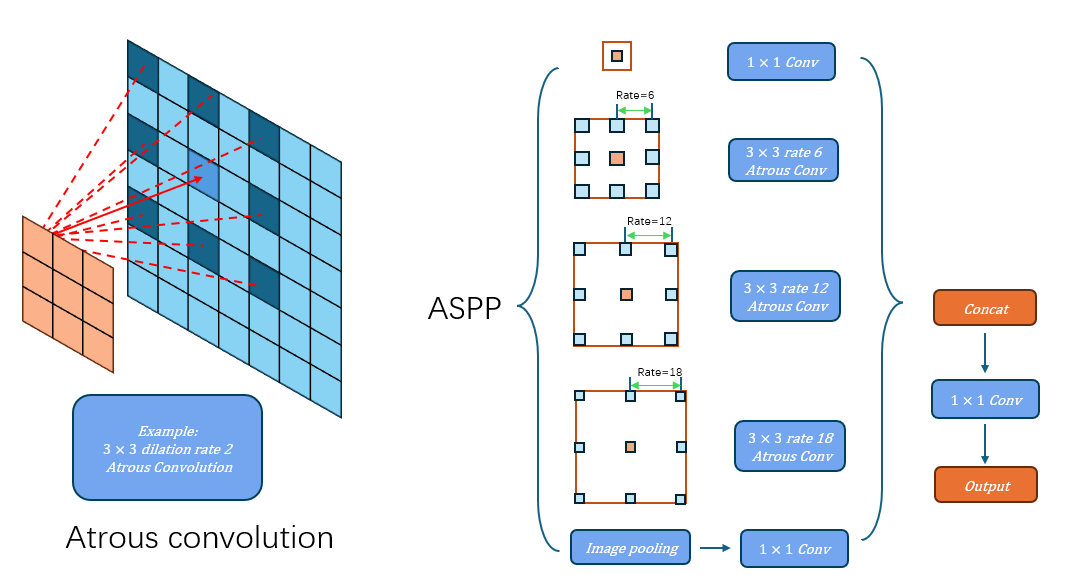
Figure 1: Diagram of atrous convolution and atrous spatial pyramid pooling (ASPP). Atrous convolution captures target features at various scales by using dilation rates that introduce controlled intervals between kernel elements. ASPP integrates several parallel atrous convolutions with different dilation rates, along with a max-pooling layer. The outputs from these layers are then merged and refined using a 1×1 convolution to produce the final feature map.
3.2. Deeplabv3+ structure
DeepLabv3+ Structure: Figure 2 gives an overview of the structure of DeepLabv3+, a powerful architecture for semantic segmentation tasks that builds upon the strengths of both deep convolutional networks and spatial pyramid pooling techniques. The network consists of an encoder and a decoder.
The encoder extracts features from the input image using a sequence of convolutional layers. Leveraging deep convolutional neural networks (DCNN), such as ResNet or Xception, The encoder conducts downsampling operations that gradually decrease the spatial resolution of the feature maps while capturing progressively more abstract semantic information. [8, 9]. It provides high-resolution and low-resolution features to the decoder for further operations. In this experiment, both low-level and high-level features are extracted from the input images by the ResNet backbone. The high-level features are then processed through Atrous Spatial Pyramid Pooling (ASPP) to effectively capture multi-scale information crucial for accurate segmentation. The resulting features are concatenated and further refined using 1×1 convolutions, yielding feature maps enriched with essential contextual details for precise segmentation.
The decoder in DeepLabv3+ upscales the low-resolution feature maps generated by the encoder to recover the spatial dimensions of the original input. The decoder generates a dense pixel-wise prediction map, where each pixel is assigned a class label corresponding to the objects present in the image. To achieve this, the feature map output from the encoder undergoes a 4 \( × \) upsampling using bilinear interpolation. The upsampled feature map is subsequently concatenated with the corresponding low-level features extracted earlier by the encoder, after applying a 1×1 convolution to enhance dimensionality. A subsequent 3 \( × \) 3 convolution refines the features, and a final 4x upsampling restores the feature map to the original resolution, producing the final segmentation output.
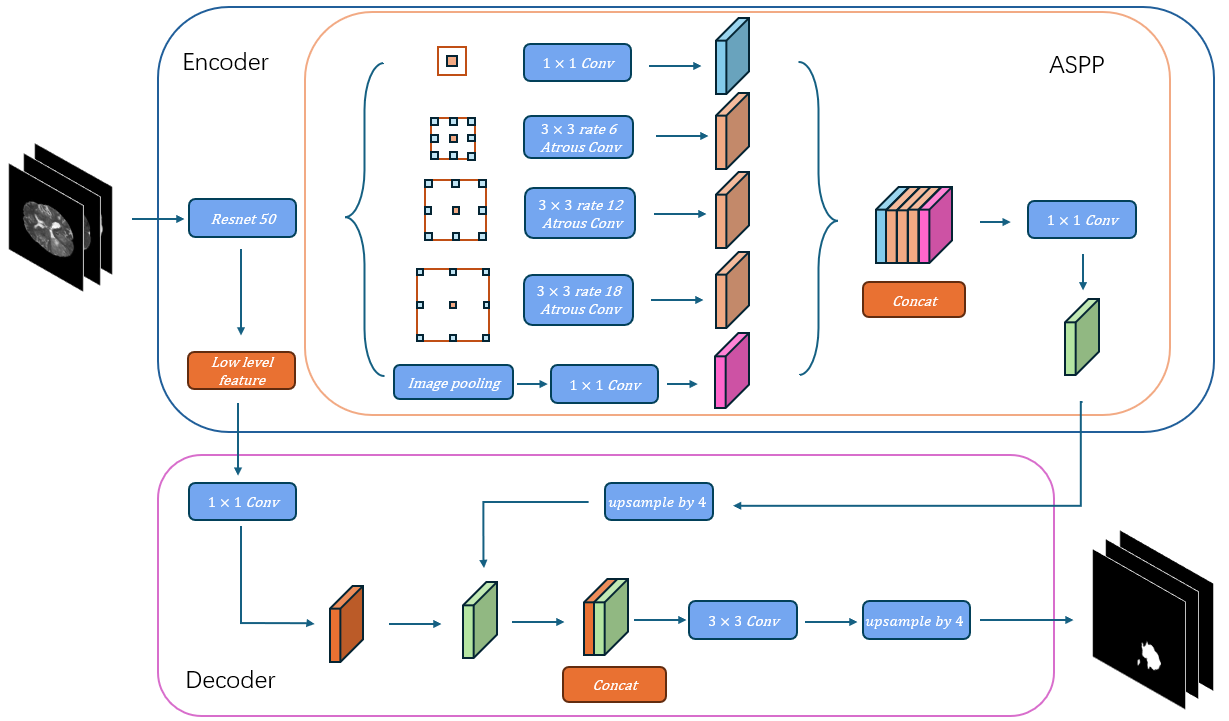
Figure 2: Schematic diagram of the DeepLabv3+ architecture used for brain tumor segmentation. The input to the model consists of three MRI modalities (T1CE, T2, and FLAIR), The backbone model ResNet is followed by an ASPP module. The output from the ASPP module is further refined using a decoder module that upsamples the feature maps to the original input resolution, producing a detailed segmentation map., and the output is a segmented map indicating tumor regions.
3.3. Dataset
The objective of this study is to assess the effectiveness of the DeepLabv3+ model in brain tumor segmentation using the BraTS2020 dataset, which comprises imaging data from real patients. This dataset includes 369 samples, each with four distinct MRI modalities: T1, T1CE, T2, and FLAIR. Since the pre-trained DeepLabv3+ model is designed to accept three-channel RGB inputs, we selected the T1CE, T2, and FLAIR modalities as the input channels for our model. The input images were resampled to a resolution of 240 x 240 pixels, maintaining a three-channel structure. The corresponding segmentation label were utilized as the ground truth for model training and evaluation.
4. Experimental Setup
4.1. Data Preprocessing
The input data consists of T1CE, T2, and FLAIR modalities, resized to 240x240 pixels. The images were normalized by subtracting the dataset's mean and dividing by its standard deviation.
4.2. Model Architecture
We utilized the DeepLabv3+ architecture with a ResNet-50 backbone pre-trained on ImageNet. The model was adapted for multi-class brain tumor segmentation with three input channels and one output segmentation map.
4.3. Optimization and Learning Rate Strategy
The AdamW optimizer with an initial learning rate of 0.001 was applied to the model. To ensure efficient training and prevent overfitting, we employed a polynomial learning rates strategy, where the learning rate gradually decreases according to the following function:
\( lr={base_{lr}}×{( \frac{1-epoch}{{num_{epoch}}})^{power}}\ \ \ (2) \)
Where the lr denotes the updated learning rate, base_lr refers to the base learning rate, epoch indicates the number of iterations, num_epoch is the maximum iteration limit, and power regulates the curve's shape. In this study, the power is set to 0.9 for maintaining a high learning rate at the start of training and for avoiding local minima.
4.4. Training Procedure
The training procedure was conducted on the BraTS2020 dataset, with 315 training samples, 17 validation samples, and 37 test samples. The model was trained for a total of 100 epochs using a batch size of 16. Early stopping was implemented using the validation Dice score, allowing for a patience of 10 epochs to avoid overfitting.
4.5. Evaluation Metrics
In this study, the evaluation of the model's performance was conducted using the Dice Coefficient. However, Dice Loss alone may not be the best choice for the loss function. Although Dice Loss is effective in handling class imbalance, it may introduce gradient instability during backpropagation, which can impair the model's convergence and overall training stability. To address these issues, we employed a hybrid loss function that integrates the Dice Coefficient and Cross-Entropy Loss. This combined approach capitalizes on the ability of loss function to accurately segment small regions while leveraging the smoothing properties of Cross-Entropy Loss to stabilize the training process. The composite loss function is defined as follows:
\( Dice Cofficient(A,B)=\frac{2|A∩B|}{|A|+|B|}\ \ \ (3) \)
Here, A represents the pixels in the predicted segmentation, while B denotes the set of pixels in the ground truth segmentation. Usually, the denominator contains a small number to prevent the value from being 0.
\( Cross Entropy Loss=\frac{1}{N}\sum _{i}{L_{i}}=\frac{1}{N}\sum _{i}\sum _{c=1}^{M}{y_{ic}}log{({p_{ic}})}\ \ \ (4) \)
where M is the number of classes, \( {y_{ic}} \) is an indicator function that equals 1 if the element belongs to class c, and \( { p_{ic}} \) represents the predicted probability that the observation belongs to class c.
\( Total Loss=Cross Entropy Loss+Dice Cofficient\ \ \ (5) \)
By combining these two loss functions, our approach aims to balance the strengths of each, enhancing both the stability of the training process and the overall segmentation accuracy of the model.
Optimizer: The AdamW optimizer was used, with a weight decay coefficient of 1e-4 to prevent overfitting. This opn odify the learning rates for each individual parameter, which is particularly useful in deep learning tasks with high-dimensional data.
Epochs: The model was trained for 200 epochs, as this number of epochs was found to be sufficient for the model to converge while avoiding overfitting.
Dropout Rate: A dropout rate of 0.1 was applied in the fully connected layers of the network. This regularization method was employed to mitigate overfitting by randomly dropping units during training, encouraging the network to learn more robust features
Evaluation of different learning rate update strategies:
This experiment also evaluates the performance of different learning rate strategies, which is shown in table 1.
Table 1. The results of employing different learning rate strategies.
Strategy | Related Parameters | Dice Coefficient |
Poly | Power = 0.9 | 0.93 |
Step | Step size = 30 | 0.87 |
Exponential Decay | New_lr = 0.9 \( × \) lr | 0.78 |
Cosine Annealing | T_Max = 50 | 0.92 |
5. Conclusion
In this study, we introduce a novel method for MRI image segmentation that leverages the deep learning principles of convolutional networks and data augmentation to make the most of the available labeled images .The DeepLabv3+ architecture utilizes an encoder-decoder structure designed to capture both low-level and high-level features. The encoder efficiently extracts spatial information, while the decoder reconstructs precise segmentation maps, enabling the model to achieve robust performance. Training was conducted over 200 epochs on a high-performance server equipped with an NVIDIA RTX 3090 GPU (24GB). The model demonstrated superior segmentation accuracy, achieving an average Dice score of 0.935 on the testing dataset. For comparison, the benchmark model, U-Net, was evaluated on the same dataset and achieved a Dice score of 0.932, underscoring the efficacy of the DeepLabv3+ architecture for MRI segmentation tasks.
However, the drawback of the model still exists. The convolution operation still processes information from each modality independently, making it unable to explicitly model the complementarity and interaction between different modalities. The convolution operation is limited by its local receptive field, which may not effectively capture long-range dependencies between modalities or fully integrate global features.
Future research will focus on exploring more advanced techniques for improving brain segmentation accuracy. Currently, most methodologies primarily focus on two-dimensional feature extraction. However, MRI data consist of 3-dimensional information with more complex relationships among neighboring layers. Evaluating inter-slice dependencies may significantly enhance segmentation performance, as 3D volumetric data capture richer spatial context. Current segmentation models that process individual slices often overlook this information, and even in models incorporating adjacent slices, the inter-slice context may still be underutilized. Addressing these limitations by integrating 3D data more effectively will be a key focus in our future work.
References
[1]. Han B, Zheng R, Zeng H, et al. Cancer incidence and mortality in China, 2022[J]. Journal of the National Cancer Center, 2024, 4(1): 47-53.
[2]. Menze B H, Jakab A, Bauer S, et al. The multimodal brain tumor image segmentation benchmark (BRATS)[J]. IEEE transactions on medical imaging, 2014, 34(10): 1993-2024.
[3]. Nawaz A, Akram U, Salam A A, et al. VGG-UNET for brain tumor segmentation and ensemble model for survival prediction[C]//2021 International Conference on Robotics and Automation in Industry (ICRAI). IEEE, 2021: 1-6.
[4]. Havaei M, Davy A, Warde-Farley D, et al. Brain tumor segmentation with deep neural networks[J]. Medical image analysis, 2017, 35: 18-31.
[5]. Iqbal S, Ghani M U, Saba T, et al. Brain tumor segmentation in multi‐spectral MRI using convolutional neural networks (CNN)[J]. Microscopy research and technique, 2018, 81(4): 419-427.
[6]. Zhao X, Wu Y, Song G, et al. A deep learning model integrating FCNNs and CRFs for brain tumor segmentation[J]. Medical image analysis, 2018, 43: 98-111.
[7]. Chen L C, Zhu Y, Papandreou G, et al. Encoder-decoder with atrous separable convolution for semantic image segmentation[C]//Proceedings of the European conference on computer vision (ECCV). 2018: 801-818.
[8]. He K, Zhang X, Ren S, et al. Deep residual learning for image recognition[C]//Proceedings of the IEEE conference on computer vision and pattern recognition. 2016: 770-778.
[9]. Chollet F. Xception: Deep learning with depthwise separable convolutions[C]//Proceedings of the IEEE conference on computer vision and pattern recognition. 2017: 1251-1258.
Cite this article
Chen,Y. (2024). Multimodal Brain Tumor Segmentation Based on Multi-Scale Feature Extraction Network. Applied and Computational Engineering,106,87-92.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 2nd International Conference on Machine Learning and Automation
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Han B, Zheng R, Zeng H, et al. Cancer incidence and mortality in China, 2022[J]. Journal of the National Cancer Center, 2024, 4(1): 47-53.
[2]. Menze B H, Jakab A, Bauer S, et al. The multimodal brain tumor image segmentation benchmark (BRATS)[J]. IEEE transactions on medical imaging, 2014, 34(10): 1993-2024.
[3]. Nawaz A, Akram U, Salam A A, et al. VGG-UNET for brain tumor segmentation and ensemble model for survival prediction[C]//2021 International Conference on Robotics and Automation in Industry (ICRAI). IEEE, 2021: 1-6.
[4]. Havaei M, Davy A, Warde-Farley D, et al. Brain tumor segmentation with deep neural networks[J]. Medical image analysis, 2017, 35: 18-31.
[5]. Iqbal S, Ghani M U, Saba T, et al. Brain tumor segmentation in multi‐spectral MRI using convolutional neural networks (CNN)[J]. Microscopy research and technique, 2018, 81(4): 419-427.
[6]. Zhao X, Wu Y, Song G, et al. A deep learning model integrating FCNNs and CRFs for brain tumor segmentation[J]. Medical image analysis, 2018, 43: 98-111.
[7]. Chen L C, Zhu Y, Papandreou G, et al. Encoder-decoder with atrous separable convolution for semantic image segmentation[C]//Proceedings of the European conference on computer vision (ECCV). 2018: 801-818.
[8]. He K, Zhang X, Ren S, et al. Deep residual learning for image recognition[C]//Proceedings of the IEEE conference on computer vision and pattern recognition. 2016: 770-778.
[9]. Chollet F. Xception: Deep learning with depthwise separable convolutions[C]//Proceedings of the IEEE conference on computer vision and pattern recognition. 2017: 1251-1258.









