1. Introduction
1.1. Importance of subsurface ecosystem
Subsurface sediments are not only significant as geologic formations beneath the Earth’s surface but also play a crucial role in environmental microbiology. Research indicates that these sediments serve as important reservoirs for microbial diversity and functional potential. Microbes within these sediments can utilize a variety of organic and inorganic substances for energy metabolism and nutrient cycling[1]. These microbial communities are not only vital to subsurface ecosystems but also have profound impacts on global biogeochemical cycles.
Further studies show that microbes in subsurface sediments can survive and thrive under extreme environmental conditions, such as high temperatures, pressures, and nutrient limitations[2]. Additionally, these microorganisms may play a key role in the cycling of subsurface methane and sulfur, affecting the release of methane into groundwater and the atmosphere[3]. Therefore, research on microbial communities in subsurface sediments is essential for understanding both the structure and function of subsurface ecosystems and their implications for global carbon, nitrogen, and sulfur cycles.
1.2. The role of underground microorganisms in subsurface ecosystem
Subsurface microorganisms occupy a unique niche within subsurface sediment ecosystems, where they play indispensable roles in maintaining the health and stability of these environments. These microorganisms, often thriving in extreme and isolated conditions, are the engines driving biogeochemical cycling and overall ecosystem dynamics.
As emphasized by Orcutt et al.[2], subsurface microbes exhibit remarkable adaptability, flourishing in challenging environments characterized by high temperatures, intense pressures, and limited nutrient availability. Their resilience and metabolic versatility enable them to utilize a wide range of organic and inorganic compounds, facilitating crucial nutrient cycling processes such as carbon fixation, nitrogen fixation, and sulfur oxidation-reduction reactions.
The significance of these microorganisms extends beyond the confines of the subsurface. Their metabolic activities, particularly those involved in methane production and consumption, as well as sulfur cycling, have profound impacts on global biogeochemical cycles[3]. For instance, methane-producing microbes (methanogens) contribute to the global methane budget, a potent greenhouse gas, while sulfate-reducing bacteria play a critical role in the sulfur cycle, influencing the solubility and mobility of sulfur compounds in the subsurface.
Moreover, the diversity and functional capabilities of subsurface microorganisms hold the key to understanding the complex interactions within these ecosystems. Their roles in nutrient cycling, energy transformation, and the degradation of organic matter are integral to maintaining the balance and productivity of subsurface environments.
In summary, subsurface microorganisms constitute a vital component of subsurface sediment ecosystems, shaping their biogeochemical characteristics and influencing global cycles. Elucidating their diversity, metabolic capabilities, and ecological niches is essential for a comprehensive understanding of subsurface ecology and its broader implications for the Earth system.
1.3. Research objectives and questions
The purpose of this study is to explore the distribution characteristics and ecological functions of underground microorganisms in the subsurface sediment ecosystem by analyzing the microbial community structure in the subsurface sediment cores in Tennessee, USA. The main objectives of the study include:
To reveal the diversity and abundance distribution of microorganisms in subsurface sediments.
Determine the taxonomic characteristics of microbial communities in different sedimentary layers.
To evaluate the relationship between microbial community structure and physicochemical properties of sediments.
To investigate the possible ecological functions and biogeochemical cycles of subsurface microorganisms in surface ecosystems.
2. Materials and methods
2.1. Sample collection and procession
Sediment core samples were meticulously gathered from a Tennessee location in the United States, employing sterile methods to preserve the microbial ecosystems present across the sediment strata. The core, spanning five meters in length, was meticulously divided into six discrete strata, each corresponding to a precise depth bracket: 0.1-0.9 meters, 0.9-1.5 meters, 1.5-2.4 meters, 2.4-3.0 meters, 3.0-4.0 meters, and 4.0-4.6 meters. These strata were designated as "FRC306_01" to "FRC306_06" for uniformity in sample identification. Subsequently, the samples were hermetically sealed within airtight containers and conveyed to the laboratory under stable temperature conditions to avert any disruption to the microbial communities that might arise from environmental fluctuations during transportation [1].
2.2. Microbial DNA extraction and sequencing
DNA extraction from the sediment samples was performed using commercially available kits, following the manufacturer's protocols. This process involved cell lysis, DNA purification, and quantification. The quality and integrity of the extracted DNA were assessed using spectrophotometry and agarose gel electrophoresis, which are standard methods in molecular biology for evaluating DNA concentration and purity [2].
Amplicon libraries targeting the 16S rRNA gene were constructed from the purified DNA. The 16S rRNA genes from bacteria and archaea were amplified using PCR with specific primers designed to target these genes. The amplicons were then purified, quantified, and sequenced using Illumina's high-throughput sequencing platform, which allows for the simultaneous analysis of a large number of DNA sequences, providing a comprehensive view of the microbial community composition [3].
2.3. Data analysis methods
The original sequencing data undergo quality control steps, including removal of low-quality sequences, host contamination, and chimeras, to ensure the accuracy of subsequent analyses. High-quality sequence data is manipulated through bioinformatics software such as QIIME2 or Mothur.
Taxonomic analysis: Taxonomic annotation of 16S rRNA gene sequences using the Ribosomal Database Project (RDP) or SILVA database. Sequences were assigned to corresponding bacterial or archaea groups by similarity search.
Community structure analysis: Using methods such as principal component analysis (PCA) or non-metric multidimensional scaling analysis (NMDS) to explore differences in microbial community structure between different samples.
Association analysis of environmental factors: Correlation analysis of microbial community data with physical and chemical properties of sediments (such as pH, organic matter content, nitrogen content, etc.) to assess the impact of environmental factors on microbial community structure.
Taxonomic Analysis: The 16S rRNA gene sequences were annotated using the Ribosomal Database Project (RDP) or SILVA database, which are comprehensive resources for microbial taxonomy based on 16S rRNA gene sequences. These sequences were compared to the databases to assign them to their corresponding bacterial or archaeal groups [5].
Abundance Statistics: The relative abundance of different microbial groups in each sample was estimated using sequence counts obtained from sequencing. Statistical software such as R or Python was used for data visualization, which allowed for the graphical representation of the abundance distribution of microbial communities across different depth layers [6].
Community Structure Analysis: Multivariate statistical methods, such as principal component analysis (PCA) or non-metric multidimensional scaling (NMDS), were employed to explore differences in microbial community structure between samples [7].
Association Analysis of Environmental Factors: Correlation analysis was performed between microbial community data and the physical and chemical properties of sediments, including pH, organic matter content, and nitrogen content. This analysis aimed to assess the impact of environmental factors on microbial community structure and to identify key factors influencing community composition [8].
3. Results
3.1. Microbial community structure of the sediment cores
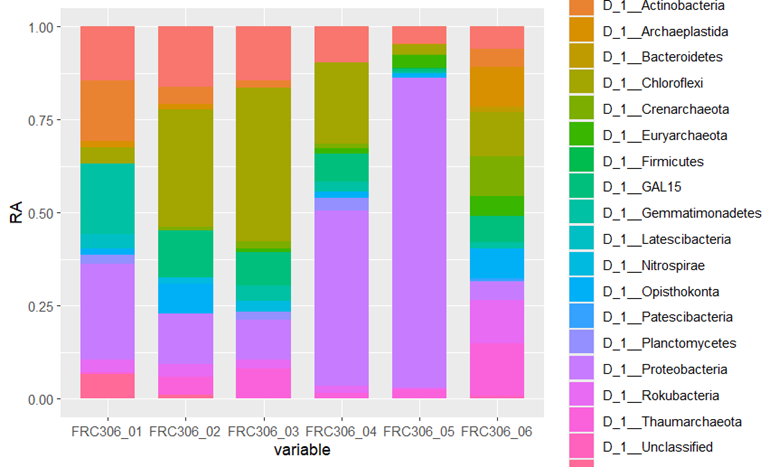
Figure 1: Relative abundances of microorganisms at the phylum level
The microbial community of the sediment core shows a distinct vertical stratification structure, which is closely related to the physicochemical gradient of the sediment. At shallower layers, such as 0.1-0.9 m depth, the microbial communities may be dominated by aerobic bacteria, which are involved in the initial decomposition process of organic matter. As depths increase, such as 0.9-1.5 m and 1.5-2.4 m, the oxygen concentration in the sediment decreases and anaerobic bacteria begin to dominate, carrying out processes such as sulfate reduction or methanogenesis. At deeper levels, such as 2.4-3.0 m, 3.0-4.0 m, and 4.0-4.6 m, the microbial community may consist of specific anaerobic bacteria that are adapted to extreme anaerobic conditions.
Microbial communities in sediment cores are composed of a variety of phyla, including but not limited to Acidobacteria, Chloroflexi, Proteobacteria, Actinobacteria, and Crenarchaeota and Euryarchaeota in Archaea (Figure 1). These microorganisms play different roles in the ecosystem, such as the decomposition of organic matter, nitrogen cycle, sulfur cycle and other key ecological processes.
In such a network map(Figure 3), we can infer the diversity and complexity of the microbial community. For example, clusters of green nodes may represent microbial communities in the 3.0 to 4.0 million base pairs (mbs), while clusters of orange nodes may represent communities in the 4.0 to 6.0 mbs range. These color-coded clusters show a high degree of correlation within the microbiome, as well as the interactions between the different communities.
3.2. The distribution of microbial abundance across different sedimentary layers
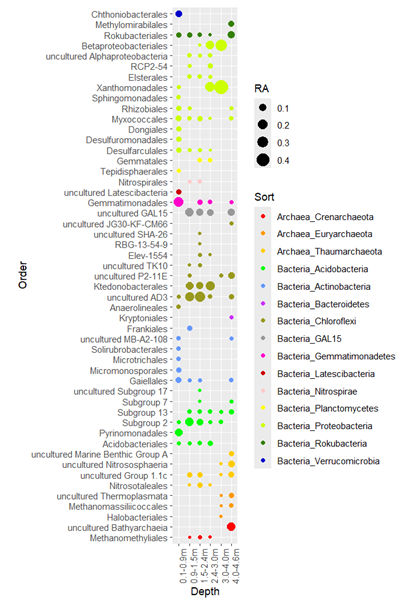
Figure 2: Relative abundances of microorganisms at the order level atdifferent depths
This chart (Figure 2) shows the relative abundance of various microbial classes at different depths. The X-axis of the chart labels multiple specific classes of organisms, each of which is represented by a different color. The Y-axis represents depth, ranging from 0-1 mbs to 4.0-4.65 mbs. The colored dots are distributed in different sizes along these axes, with the size of each dot representing the relative abundance of the category at a particular depth. There are legends of categories and relative abundance at the bottom of the chart.
In our analysis of sediment cores at different depths, we found vertical patterns of microbial abundance and diversity. Microbial abundance was generally higher in the shallower sediments, which may be related to the higher organic matter content and oxygen penetration in these horizons. Organic matter is an important source of energy for microbial growth, while oxygen is necessary for the survival of many aerobic microorganisms. Therefore, the improvement of these conditions is conducive to the reproduction and activity of microorganisms.
In contrast, the abundance of microbes was significantly reduced in the deeper sediments. This may be due to the lower availability of nutrients in the deeper sediments, as well as the lack of oxygen. Under these conditions, microorganisms may need to rely on other energy sources, such as nitrates, sulfates, or iron ions, for anaerobic metabolism. In addition, microorganisms in deep sediments may have longer generation times, resulting in their relatively low abundance.
It should be highlighted that particular microbial communities, including Acidobacteria, exhibit a significant presence within distinct sedimentary strata. This occurrence likely underscores the adeptness of these microorganisms to particular environmental niches, such as their resilience to acidic conditions or nutrient-depleted settings. These microscopic entities likely fulfill crucial ecological roles within the profundity of sediments, engaging in the sluggish degradation of organic matter or conducting energy metabolism in oxygen-deprived environments.
3.3. Relationships between microbial communities and environmental factors
Further analysis revealed a correlation between microbial community structure and the geochemistry of sediments. For example, pH was significantly positively correlated with the abundance of certain bacterial groups, while organic matter content was correlated with the abundance of archaea communities. In addition, microorganisms from the phylum Nitrospira increased in abundance in sedimentary layers with high nitrate and nitrite content, suggesting that they may play an important role in the nitrogen cycle process. Through multiple regression analysis, we identified key environmental factors that influence the structure of microbial communities and explored how together they shaped the microbial ecosystem in cavern sediments.
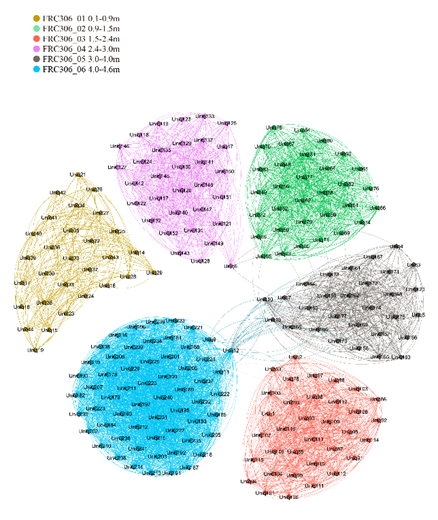
Figure 3: Map of the co-occurrence network
4. Discussion
4.1. Ecological significance of stratified distribution of microbial communities
The microbial communities in the subsurface sediment cores show obvious stratified distribution, which may be closely related to the material circulation and energy flow in the sediments. In the shallower sediments, microbial diversity and abundance are higher, which may be related to more input of organic matter, higher oxygen penetration and temperature change. These conditions promoted the metabolic activities of microorganisms, which in turn affected the functions and services of the cave ecosystem. In contrast, the microbial communities in the deep sediments may have adapted to the low-energy environment, and they may have been involved in slow organic matter degradation processes, such as anaerobic respiration and fermentation[11].
4.2. The function of subsurface microorganisms in subsurface ecosystems
Underground microorganisms play a variety of roles in subsurface ecosystems. By breaking down organic matter, they provide energy and nutrients to the ecosystem and support the foundation[12]of the subsurface biome. In addition, certain microbial groups such as Nitrospira play a key role in the nitrogen cycle by oxidizing nitrite to produce nitrates, which in turn affect nutrient access to plants and the chemical composition of groundwater. These microbial processes not only maintain the stability of subsurface ecosystems, but also have an impact on surface ecosystems and human activities.
4.3. Mechanisms of interaction between microbial communities and subsurface environment
The structure and function of the microbial community are affected by various physical and chemical factors in the subsurface environment. For example, the pH value, organic matter content, water status and mineral composition of sediments may affect the distribution and activity[13] of microorganisms. Through correlation analysis and multiple regression models, we were able to identify key environmental factors that influence microbial community structure and speculate on how they shape community structure by influencing microbial metabolic pathways. In addition, interactions between microbial communities, such as symbiosis, competition, and predation relationships, may also influence their niches and evolution.
4.4. Environmental Adaptation of Subsurface Microorganisms
The environmental adaptation of subsurface microbial communities is crucial for their survival and reproduction, as these microscopic organisms inhabit a world far removed from the surface, where conditions can be harsh and unpredictable. Studies have shown that subsurface microorganisms possess remarkable capabilities to adapt to various environmental stresses, such as extreme temperatures ranging from the scorching heat of hydrothermal vents to the freezing cold of polar ice caps, immense pressures that would crush most surface life forms, and nutrient limitations that challenge their ability to obtain the essential elements required for growth and metabolism. These adaptations are fundamental for their roles in subsurface ecosystems, where they play pivotal roles in nutrient cycling, biodegradation, and even in the formation of mineral deposits [14].
For instance, consider the thermophilic bacteria that thrive in the hot springs of Yellowstone National Park. These microorganisms have evolved to not only survive but to flourish in water temperatures that exceed the boiling point. Their cellular machinery, including enzymes and proteins, is adapted to function optimally at these high temperatures, a feature that has significant implications for biotechnological applications, such as the development of heat-stable enzymes for industrial processes.
Similarly, piezophilic (pressure-loving) microbes, found in the deep ocean trenches, have adapted to withstand pressures exceeding 1,000 atmospheres. These organisms have unique cell membrane compositions and protein structures that prevent them from being crushed by the immense pressure. Their existence challenges our understanding of life's limits and provides insights into the potential for life on other planets or moons with similarly extreme conditions.
Nutrient limitations are another environmental stress that subsurface microbes have mastered. In environments where essential nutrients like nitrogen, phosphorus, and sulfur are scarce, these microbes have developed efficient strategies for nutrient acquisition and recycling. Some can fix atmospheric nitrogen into a usable form, while others can solubilize and utilize phosphorus from minerals. These adaptations are not only crucial for the survival of the microbes themselves but also for the broader ecosystem, as they contribute to the cycling of these vital elements.
Moreover, the ability of subsurface microbes to form biofilms and consortia allows them to survive in environments where individual cells might not. These communities can share resources, provide mutual protection, and even communicate through chemical signals, coordinating their activities for the benefit of the group. This social behavior is particularly important in nutrient-poor or otherwise challenging environments, where cooperation can mean the difference between life and death.
In conclusion, the environmental adaptation of subsurface microbial communities is a testament to the resilience and versatility of life. These adaptations are not only crucial for the survival and reproduction of the microbes themselves but also play a critical role in maintaining the health and function of subsurface ecosystems. As our understanding of these hidden worlds deepens, it becomes increasingly clear that subsurface microbes are not just survivors but active participants in shaping the conditions of their environment, with implications that extend far beyond their immediate surroundings [14].
4.5. Metabolic Functions of Subsurface Microbial Communities
Subsurface microorganisms play a significant role in biogeochemical cycles. They are involved in the decomposition of organic matter, nutrient cycling, and the production and consumption of greenhouse gases, impacting global carbon, nitrogen, and sulfur cycles [14].
4.6. Biomarkers of Subsurface Microbial Communities
The structure and function of subsurface microbial communities can be characterized by biomarkers. These biomarkers can help us understand the ecological functions and environmental responses of subsurface microorganisms [15].
4.7. Conservation and Management of Subsurface Microbial Communities
The conservation and management of subsurface microbial communities are essential for maintaining the health and stability of subsurface ecosystems. This requires a comprehensive consideration of the diversity, ecological functions, and environmental responses of subsurface microbial communities [17].
Microbial communities in the subsurface environment, which include bacteria, archaea, fungi, and other microorganisms, play a crucial role in nutrient cycling, biodegradation, and the maintenance of soil structure. These microorganisms can break down organic matter, recycle nutrients, and even influence the chemical composition of groundwater. For instance, certain bacteria can convert toxic heavy metals into less harmful forms, while others are involved in the nitrogen cycle, transforming nitrogen into forms that plants can use.
The diversity of these microbial communities is vast and complex. Each microbe species contributes uniquely to the ecosystem, and their interactions can be intricate and interdependent. For example, some bacteria form symbiotic relationships with plant roots, aiding in nutrient absorption while receiving carbohydrates in return[18]. This intricate web of life is sensitive to changes in the environment, such as alterations in pH, temperature, or the introduction of pollutants.
Understanding the ecological functions of these communities is vital for their conservation. Microbes can act as indicators of environmental health, as changes in their populations or activities can signal disturbances in the ecosystem. For example, a decline in the population of certain bacteria that are efficient at breaking down oil could indicate that an oil spill has occurred, and these microbes are being overwhelmed.
Environmental responses of subsurface microbial communities to changes such as climate change, pollution, and human activities must also be considered. These responses can range from shifts in microbial populations to changes in metabolic activities. For example, increased temperatures due to global warming could lead to the proliferation of thermophilic microbes, potentially altering the nutrient dynamics of the ecosystem.
Conservation efforts must be multifaceted, involving the protection of habitats, the prevention of pollution, and the restoration of disturbed areas[18]. Management practices should be informed by scientific research that monitors microbial diversity and function. For example, land-use planning can incorporate buffer zones to protect natural microbial habitats from agricultural runoff or urban development.
In addition, bioremediation techniques can be employed to harness the natural abilities of microbes to clean up contaminated sites[19]. This involves stimulating microbial activity through the addition of nutrients or oxygen, or by adjusting environmental conditions to favor the growth of microorganisms that can break down pollutants.
Education and public awareness are also key components of conservation and management strategies. By informing the public about the importance of subsurface microbial communities, we can foster a greater appreciation for these unseen ecosystems and encourage responsible behavior that minimizes negative impacts.
In conclusion, the conservation and management of subsurface microbial communities are critical for the sustainability of our planet's subsurface ecosystems. By understanding and protecting these communities, we can ensure that they continue to provide essential services that support life on Earth[20]. Future research should focus on developing innovative approaches to monitor, protect, and restore these vital microbial communities, thereby contributing to the overall health of our environment.
5. Conclusion
5.1. Main fundings of the study
Microbial communities showed significant stratification in different depth layers of subsurface sediments, which may be related to the available organic matter and the vertical distribution of electron acceptors in the sediments.
The diversity and abundance of microbial communities are strongly influenced by the geochemistry of sediments, especially environmental factors such as organic matter content, pH and nitrate concentration.
Subsurface microorganisms play a variety of potential ecological roles in subsurface ecosystems, including decomposers of organic matter, agents of nutrient cycling, and shapers of cave morphology.
These findings provide a new perspective for understanding the role of subsurface microorganisms in subsurface ecosystems and lay a foundation for further research.
5.2. Implications for the conservation and management of subsurface ecosystems
The findings of this study have important implications for the conservation and management of subsurface ecosystems:
Conservation strategies for karst cave ecosystems should take into account the complexity of subsurface microbial communities and their responses to environmental changes, especially in the face of global climate change and human disturbances.
In order to maintain the health and stability of cave ecosystems, it is necessary to protect and restore the physical and chemical properties of sediments, which are essential for the survival and functioning of subsurface microorganisms.
Future conservation efforts should include monitoring of microbial community structure and function in sediment cores, which can provide important information for assessing the health of subsurface ecosystems.
5.3. Future research direction
This study reveals the basic characteristics of subsurface microbial communities, but also points out potential directions for future research:
Further studies are needed on the metabolic functions of subsurface microbial communities and how they influence the ecological processes of subsurface ecosystems by participating in different biogeochemical cycles.
Using advanced technologies such as metagenomics and single-cell genomics, it is possible to gain a more complete understanding of the genetic diversity and potential functions of subsurface microbes, especially for those microbial taxa that have not yet been cultured.
Explore the interactions between subsurface microbial communities and subsurface biomes, and how these interactions work together to influence the structure and function of subsurface ecosystems.
Comparative analysis of microbial communities in different geographic regions and different types of subsurface ecosystems will help reveal the geographic distribution patterns and environmental adaptability of microbial communities.
In conclusion, this study not only improves our understanding of subsurface microbial communities in subsurface, but also provides an important scientific basis for the protection and sustainable management of subsurface ecosystems. Future studies will continue to deepen our understanding of subsurface microbial ecology and provide more support for the conservation of subsurface ecosystems worldwide.
References
[1]. A. Schippers, L. N. Neretin, J. Kallmeyer, T. G. Ferdelman, B. A. Cragg, R. J. Parkes, and B. B. Jorgensen. 2005. Prokaryotic cells of the deep sub-seafloor biosphere identified as living bacteria. Nature 433, 7028, https://doi.org/ 10.1038/nature03302
[2]. B. N. Orcutt, J. B. Sylvan, N. J. Knab, and K. J. Edwards. 2011. Microbial Ecology of the Dark Ocean above, at, and below the Seafloor. Microbiol. Mol. Biol. Rev. 75, 2, https://doi.org/ 10.1128/mmbr.00039-10
[3]. F. Inagaki, K. Takai, K. H. Nealson, and K. Horikoshi. 2004. <i>Sulfurovum lithotrophicum</i> gen. nov., sp nov., a novel sulfur-oxidizing chemolithoautotroph within the ε-<i>Proteobacteria</i> isolated from Okinawa Trough hydrothermal sediments. Int. J. Syst. Evol. Microbiol. 54https://doi.org/ 10.1099/ijs.0.03042-0
[4]. Adams RI, Miletto M, Taylor JW, Bruns TD (2013) Dispersal in microbes: Fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. The ISME Journal, 7, 1262-1273. DOI: 10.1038/ismej.2013.28
[5]. Adriaenssens EM, Kramer R, van Goethem MW, Makhalanyane TP, Hogg I, Cowan DA (2017) Environmental drivers of viral community composition in Antarctic soils identified by viromics. Microbiome, 5, 83. DOI: 10.1186/s40168-017-0301-7
[6]. Aguilar-Trigueros CA, Powell JR, Anderson IC, Antonovics J, Rillig MC (2014) Ecological understanding of root-infecting fungi using trait-based approaches. Trends in Plant Science, 19, 432-438. DOI: 10.1016/j.tplants.2014.02.006
[7]. Gao C, Guo LD (2013) Distribution pattern and maintenance of ectomycorrhizal fungus diversity. Biodiversity Science, 21, 488-498. (in Chinese with English abstract) DOI: 10.3724/SP.J.1003.2013.11055
[8]. Li D, Ni HW, Jiao S, Lu YH, Zhou JZ, Sun B, Liang YT (2021) Coexistence patterns of soil methanogens are closely tied to methane generation and community assembly in rice paddies. Microbiome, 9, 20. DOI: 10.1186/s40168-020-00978-8
[9]. Liu L, Barberán A, Gao C, Zhang ZC, Wang M, Wurzburger N, Wang X, Zhang R, Li JX, Zhang J (2022) Impact of urbanization on soil microbial diversity and composition in the megacity of Shanghai. Land Degradation and Development, 33, 282-293. DOI: 10.1002/ldr.4145
[10]. Liu WX, Jiang L, Yang S, Wang Z, Tian R, Peng ZY, Chen YL, Zhang XX, Kuang JL, Ling N, Wang SP, Liu LL (2020) Critical transition of soil bacterial diversity and composition triggered by nitrogen enrichment. Ecology, 101, e03053.
[11]. LIU Yubing, CAO Chunxin, HUANG Hongming, LU Huabing, LYU Xuegao, and LIU Xinhua. 2023. Effect of degradation membrane on community structure of sorghum soil microorganisms. Journal of Shanxi Agricultural University(Natural Science Edition) https://doi.org/ 10.13842/j.cnki.issn1671-8151.202311035
[12]. WANG Fangzhou, WEI Yanfeng, ZHU Fangfang, Gou Min, and TANG Yueqing. 2024. STUDY ON THE ENRICHMENT AND STABILITY OF ENDOGENOUS MICROBIAL COMMUNITY IN CRUDE OIL PHASE OF RESERVOIR PRODUCED FLUID. Environmental Engineering
[13]. Wu Weiwei, Han Xue, Wang Jiming, Sun Nianxi, and Li Yong. 2024. Alterations with Plantation Years of Fructus aurantii on Rhizosphere Microbiome and Soil Properties. Acta Pedologica Sinica
[14]. Ning, D., Wang, Y., Fan, Y., Wang, J., Van Nostrand, J. D., Wu, L., ... & Zhou, J. (2024). Environmental stress mediates groundwater microbial community assembly. Nature Microbiology. https://doi.org/10.1038/s41564-023-01573-x
[15]. Lekberg, Y., Arnillas, C. A., Borer, E. T., Bullington, L. S., Fierer, N., Kennedy, P. G., Leff, J. W., Luis, A. D., Seabloom, E. W., & Henning, J. A. (2021). Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proceedings of the National Academy of Sciences, USA, 112, 10967-10972.
[16]. Li, D., Ni, H. W., Jiao, S., Lu, Y. H., Zhou, J. Z., Sun, B., & Liang, Y. T. (2021). Coexistence patterns of soil methanogens are closely tied to methane generation and community assembly in rice paddies. Microbiome, 9, 20. https://doi.org/10.1186/s40168-020-00978-8
[17]. Abriouel, H., Franz, C. M. A. P., Ben Omar, N., et al. (2011). Diversity and Applications of Bacillus Bacteriocins. FEMS Microbiology Reviews, 35(1), 201-232. https://doi.org/10.1111/j.1574-6976.2010.00244.x
[18]. Zhou LW, Dal Yc (2013) Chinese palypore dersite: speces, mycota and ecologlcal functions Biodiversity Science, 21, 499-506. (In Chinese with English abstract
[19]. Su JQ, Huang FY, Zhu yG (2013) Antibiotic rsistancte genes in the environment. Biodiversity Science, 21, 481-487. (in Chinese with English abstract)
[20]. Kong WD (2013) A review of microblal diversty In polar terrestrial environments. Biodiversity Science, 21, 456-467. (In Chinese with English abstract)
Cite this article
Fang,J. (2025). Investigation of the Microbial Community Structures and Function in subsurface Sediments. Applied and Computational Engineering,130,119-129.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 5th International Conference on Materials Chemistry and Environmental Engineering
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. A. Schippers, L. N. Neretin, J. Kallmeyer, T. G. Ferdelman, B. A. Cragg, R. J. Parkes, and B. B. Jorgensen. 2005. Prokaryotic cells of the deep sub-seafloor biosphere identified as living bacteria. Nature 433, 7028, https://doi.org/ 10.1038/nature03302
[2]. B. N. Orcutt, J. B. Sylvan, N. J. Knab, and K. J. Edwards. 2011. Microbial Ecology of the Dark Ocean above, at, and below the Seafloor. Microbiol. Mol. Biol. Rev. 75, 2, https://doi.org/ 10.1128/mmbr.00039-10
[3]. F. Inagaki, K. Takai, K. H. Nealson, and K. Horikoshi. 2004. <i>Sulfurovum lithotrophicum</i> gen. nov., sp nov., a novel sulfur-oxidizing chemolithoautotroph within the ε-<i>Proteobacteria</i> isolated from Okinawa Trough hydrothermal sediments. Int. J. Syst. Evol. Microbiol. 54https://doi.org/ 10.1099/ijs.0.03042-0
[4]. Adams RI, Miletto M, Taylor JW, Bruns TD (2013) Dispersal in microbes: Fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. The ISME Journal, 7, 1262-1273. DOI: 10.1038/ismej.2013.28
[5]. Adriaenssens EM, Kramer R, van Goethem MW, Makhalanyane TP, Hogg I, Cowan DA (2017) Environmental drivers of viral community composition in Antarctic soils identified by viromics. Microbiome, 5, 83. DOI: 10.1186/s40168-017-0301-7
[6]. Aguilar-Trigueros CA, Powell JR, Anderson IC, Antonovics J, Rillig MC (2014) Ecological understanding of root-infecting fungi using trait-based approaches. Trends in Plant Science, 19, 432-438. DOI: 10.1016/j.tplants.2014.02.006
[7]. Gao C, Guo LD (2013) Distribution pattern and maintenance of ectomycorrhizal fungus diversity. Biodiversity Science, 21, 488-498. (in Chinese with English abstract) DOI: 10.3724/SP.J.1003.2013.11055
[8]. Li D, Ni HW, Jiao S, Lu YH, Zhou JZ, Sun B, Liang YT (2021) Coexistence patterns of soil methanogens are closely tied to methane generation and community assembly in rice paddies. Microbiome, 9, 20. DOI: 10.1186/s40168-020-00978-8
[9]. Liu L, Barberán A, Gao C, Zhang ZC, Wang M, Wurzburger N, Wang X, Zhang R, Li JX, Zhang J (2022) Impact of urbanization on soil microbial diversity and composition in the megacity of Shanghai. Land Degradation and Development, 33, 282-293. DOI: 10.1002/ldr.4145
[10]. Liu WX, Jiang L, Yang S, Wang Z, Tian R, Peng ZY, Chen YL, Zhang XX, Kuang JL, Ling N, Wang SP, Liu LL (2020) Critical transition of soil bacterial diversity and composition triggered by nitrogen enrichment. Ecology, 101, e03053.
[11]. LIU Yubing, CAO Chunxin, HUANG Hongming, LU Huabing, LYU Xuegao, and LIU Xinhua. 2023. Effect of degradation membrane on community structure of sorghum soil microorganisms. Journal of Shanxi Agricultural University(Natural Science Edition) https://doi.org/ 10.13842/j.cnki.issn1671-8151.202311035
[12]. WANG Fangzhou, WEI Yanfeng, ZHU Fangfang, Gou Min, and TANG Yueqing. 2024. STUDY ON THE ENRICHMENT AND STABILITY OF ENDOGENOUS MICROBIAL COMMUNITY IN CRUDE OIL PHASE OF RESERVOIR PRODUCED FLUID. Environmental Engineering
[13]. Wu Weiwei, Han Xue, Wang Jiming, Sun Nianxi, and Li Yong. 2024. Alterations with Plantation Years of Fructus aurantii on Rhizosphere Microbiome and Soil Properties. Acta Pedologica Sinica
[14]. Ning, D., Wang, Y., Fan, Y., Wang, J., Van Nostrand, J. D., Wu, L., ... & Zhou, J. (2024). Environmental stress mediates groundwater microbial community assembly. Nature Microbiology. https://doi.org/10.1038/s41564-023-01573-x
[15]. Lekberg, Y., Arnillas, C. A., Borer, E. T., Bullington, L. S., Fierer, N., Kennedy, P. G., Leff, J. W., Luis, A. D., Seabloom, E. W., & Henning, J. A. (2021). Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proceedings of the National Academy of Sciences, USA, 112, 10967-10972.
[16]. Li, D., Ni, H. W., Jiao, S., Lu, Y. H., Zhou, J. Z., Sun, B., & Liang, Y. T. (2021). Coexistence patterns of soil methanogens are closely tied to methane generation and community assembly in rice paddies. Microbiome, 9, 20. https://doi.org/10.1186/s40168-020-00978-8
[17]. Abriouel, H., Franz, C. M. A. P., Ben Omar, N., et al. (2011). Diversity and Applications of Bacillus Bacteriocins. FEMS Microbiology Reviews, 35(1), 201-232. https://doi.org/10.1111/j.1574-6976.2010.00244.x
[18]. Zhou LW, Dal Yc (2013) Chinese palypore dersite: speces, mycota and ecologlcal functions Biodiversity Science, 21, 499-506. (In Chinese with English abstract
[19]. Su JQ, Huang FY, Zhu yG (2013) Antibiotic rsistancte genes in the environment. Biodiversity Science, 21, 481-487. (in Chinese with English abstract)
[20]. Kong WD (2013) A review of microblal diversty In polar terrestrial environments. Biodiversity Science, 21, 456-467. (In Chinese with English abstract)









