1. Introduction
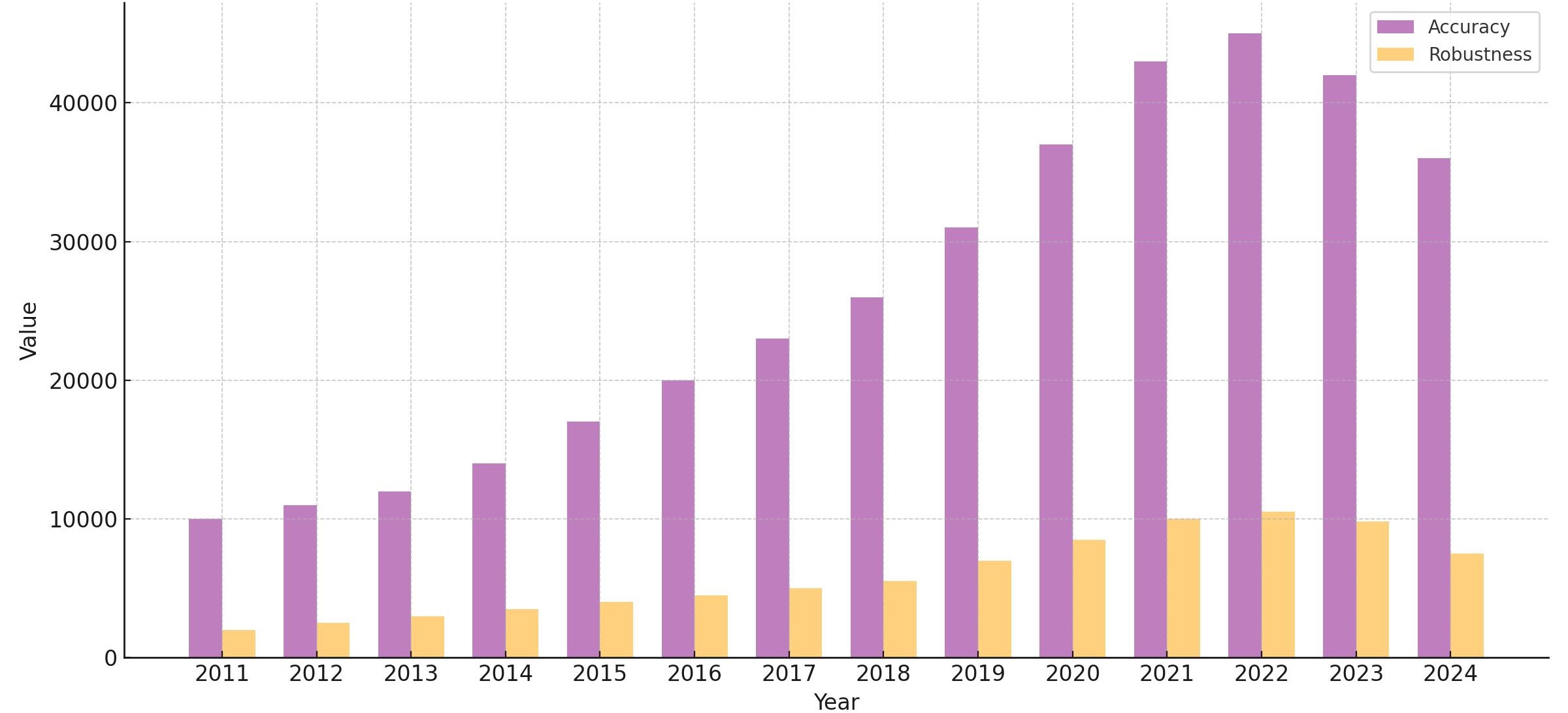
Traditionally, medical image segmentation algorithms are demonstrated by training and testing on the same dataset, which may not accurately reflect real-world scenarios where variations in data distribution arise from differences in imaging methods or devices. Additionally, a statistical analysis of research papers published over the past 14 years reveals a disproportionate focus on model accuracy compared to robustness(Figure 1). The number of papers addressing robustness is significantly lower than those focused on accuracy, highlighting a relative neglect of robustness research and underscoring the need for further investigation in this domain. To address this gap, we explore the performance of medical image segmentation algorithms when trained on one dataset and tested on another with distinguishing features. Specifically, we train the models on the Multi-Atlas Labeling Beyond the Cranial Vault (hereinafter referred to as BTCV) dataset, and evaluate their performance on the AbdomenCT-1K dataset. Both datasets consist of abdomen CT scans with multi-organ labels.
2. Methods
2.1. Datasets
2.1.1. BTCV
The BTCV dataset [2] is a publicly available medical imaging resource designed for multi-organ segmentation tasks. It comprises 50 clinically acquired CT scan images, each manually labeled for 13 abdominal organs, including the spleen, kidneys, gallbladder, esophagus, liver, stomach, aorta, inferior vena cava, portal/splenic vein, and adrenal glands.
2.1.2. AbdomenCT-1K
The AbdomenCT-1K dataset [3] is an augmented collection derived from existing single-organ datasets, incorporating four abdominal organs: the liver, kidney, spleen, and pancreas. It is a large and diverse multi-organ segmentation dataset consisting of over 1,000 CT scans of the abdomen.
2.2. Experiments
The models were selected because they all employ U-Net-like architectures but incorporate different types of basic blocks, which may affect their robustness. We designed the experimental background to fairly compare the robustness of models in the face of a large amount of unfamiliar data, utilizing a limited number of training instances under rapid training conditions. Following standardized training procedures, we set a lower tolerance level by stopping training early if the model does not improve within 25 epochs. This approach is designed to prevent overfitting and to control variables while focusing on robustness. During inference, we align the label tags of the inference instances with those in the AbdomenCT-1K dataset, while additional organ labels from the BTCV dataset are reset to 0 and treated as background. All experiments are conducted using an NVIDIA RTX 3090 GPU.
2.2.1. 3D Models
In the 3D experiments, the nnFormer [4], nnU-Net [5], and Channel-Spatial-Attention nnU-Net [6] (hereinafter referred to as CSA-nnU-Net) adhere to the adaptive architecture of nnU-Net.
2.2.2. 2D Models
In the 2D experiments, 3D data files (.nii or.nii.gz) are sliced into 2D images in a certain order and applied to the TransUNet [7] and UNet++ [8] models.
3. Results and Discussion
3.1. Experiments result
Table 1: Comparison of model performance on BTCV dataset (the average validation Dice of the saved best model) and AbdomenCT-1K dataset (the average Dice of all inferences). For each organ and the average values, the optimal Dice scores for both 3D and 2D models are highlighted in bold, while the overall best values across all five models are emphasized with a yellow background.
\resizebox{\textwidth}{!}{%
| Dice on BTCV dataset | Dice on AbdomenCT-1K dataset | |||||||||
| Label1: | Label2: | Label3: | Label4: | Average | Label1: | Label2: | Label3: | Label4: | Average | |
| Liver | Kidney | Spleen | Pancreas | Dice | Liver | Kidney | Spleen | Pancreas | Dice | |
| nnFormer | 0.9492 | 0.9005 | 0.8915 | 0.7797 | 0.8842 | 0.9629 | 0.9147 | 0.9620 | 0.7382 | 0.8945 |
| nnU-Net | 0.9455 | 0.9153 | 0.7926 | 0.7216 | 0.8506 | 0.9583 | 0.9107 | 0.9553 | 0.7874 | 0.9030 |
| CSA-nnU-Net | 0.9192 | 0.8608 | 0.8012 | 0.6257 | 0.8189 | 0.9392 | 0.9102 | 0.9387 | 0.7766 | 0.8912 |
| TransUNet | 0.9399 | 0.8045 | 0.8593 | 0.5368 | 0.7889 | 0.8482 | 0.8286 | 0.5595 | 0.3191 | 0.6388 |
| UNet++ | 0.8874 | 0.8965 | 0.6685 | 0.7800 | 0.7154 | 0.8791 | 0.7920 | 0.7452 | 0.6161 | 0.7580 |
}
3.1.1. 3D Models
As shown in Table [???], nnFormer achieved the best average Dice score in the segmentation of the liver, kidney, and spleen, while nnU-Net performed better in the pancreas. To be more specific, the maximum, minimum, and variance of Dice scores are calculated and presented in Table [???], with the best values highlighted in bold in the table. The nnFormer achieved the highest max/min Dice scores and the smallest variance across the first three classes, only slightly trailing the CSA-nnU-Net by 0.00006 in the maximum score for the kidney class. In contrast, nnU-Net held the leading position in the pancreas class. This suggests that under nearly identical training constraints, nnFormer exhibits better performance across the first three classes. Conversely, nnU-Net demonstrates better performance in the pancreas class.
Table 2: Comparison of 3D models performance on the AbdomenCT-1K dataset (including the maximum, minimum, and variance of the Dice score for each organ, with the optimal values for each item highlighted in bold).
\resizebox{\textwidth}{!}{%
| \ | Label1: Liver | Label2: Kidney | Label3: Spleen | Label4: Pancreas | ||||||||
| \ | Max | Min | Var | Max | Min | Var | Max | Min | Var | Max | Min | Var |
| nnFormer | 0.9816 | 0.8723 | 0.0003 | 0.9606 | 0.6696 | 0.0018 | 0.9916 | 0.6227 | 0.0021 | 0.9103 | 0.1583 | 0.0250 |
| nnU-Net | 0.9785 | 0.7327 | 0.0007 | 0.9590 | 0.1651 | 0.0052 | 0.9879 | 0.0838 | 0.0098 | 0.9301 | 0.3722 | 0.0106 |
| CSA-nnU-Net | 0.9701 | 0.7246 | 0.0013 | 0.9607 | 0.5987 | 0.0019 | 0.9882 | 0.0997 | 0.0152 | 0.9204 | 0.3264 | 0.0117 |
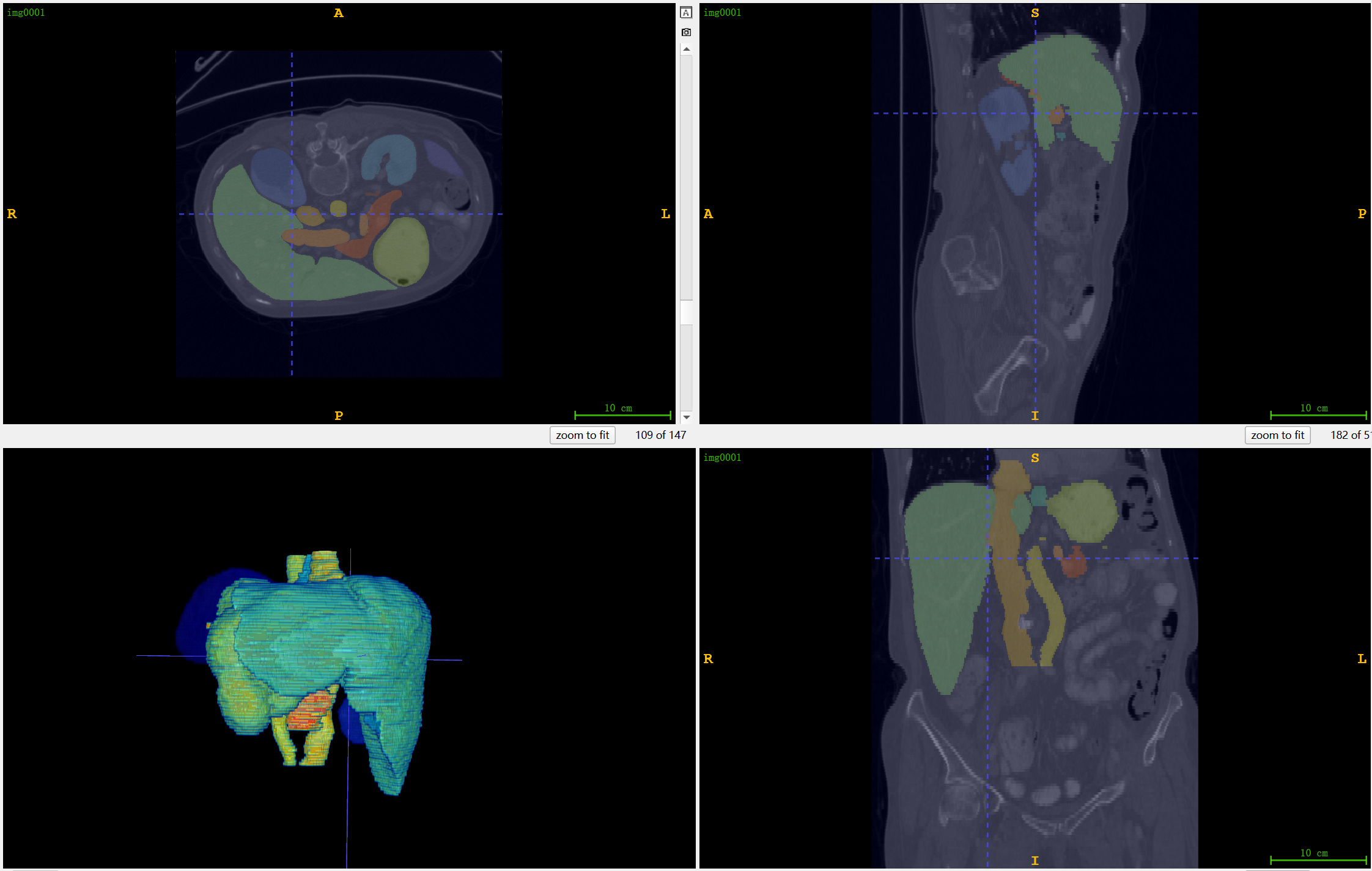
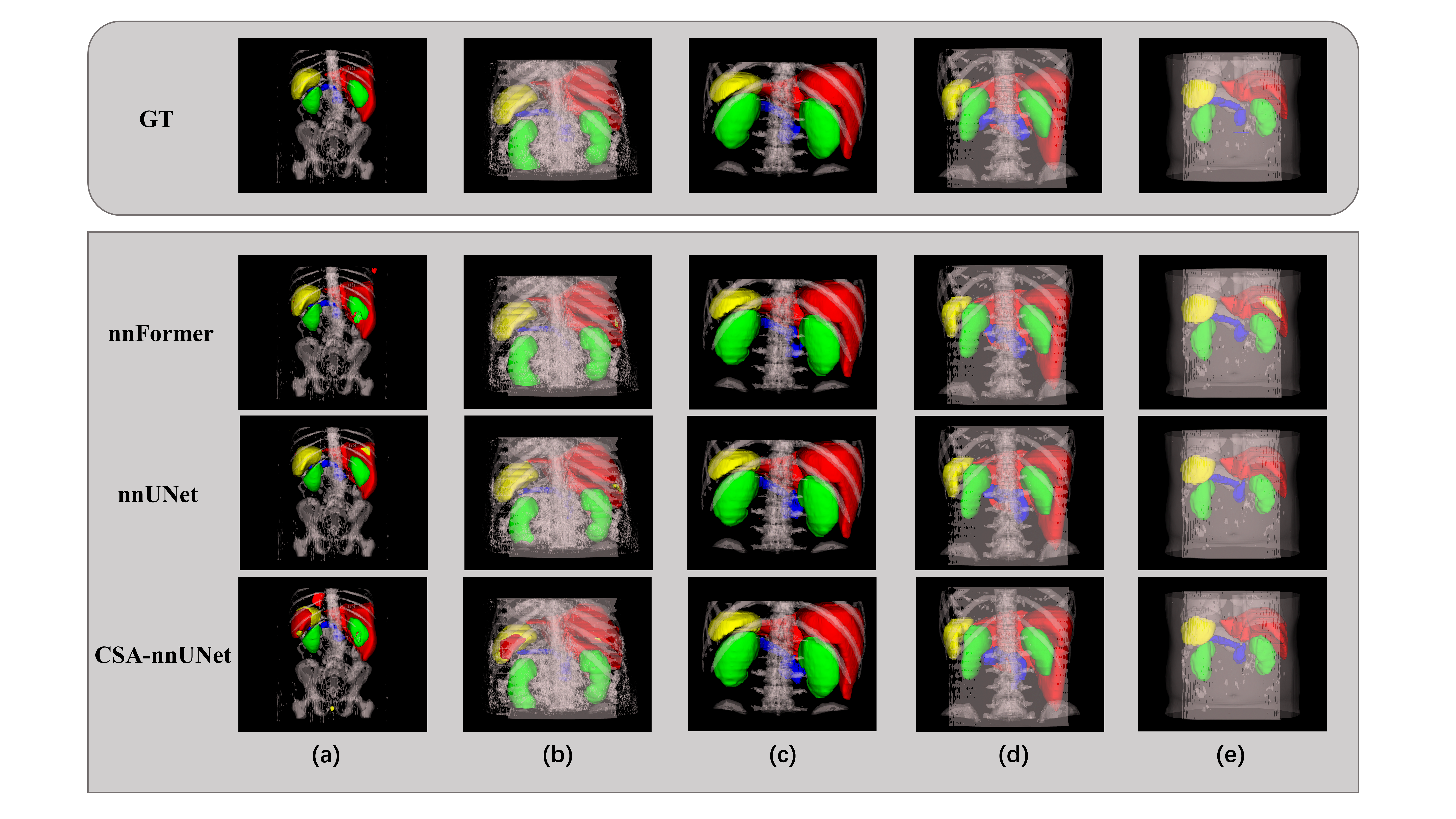
} Furthermore, we visualize [9] five instances of AbdomenCT-1K dataset and compared the ground truth and the inferences of different models, as shown in Figure 3. We also observe that the instance with the lowest Dice score was the same for both nnU-Net and CSA-nnU-Net, so we visualize and compare it in Figure 4 case (a). The poor performance in this instance is due to the misidentification of the spleen as the liver by the nnU-Net-based models. Meanwhile, the worst segmentaion of nnFormer is also presented in Figure 4 case (b), indicating the omission of pancreas and misidentification of kidney.

3.1.2. 2D Models
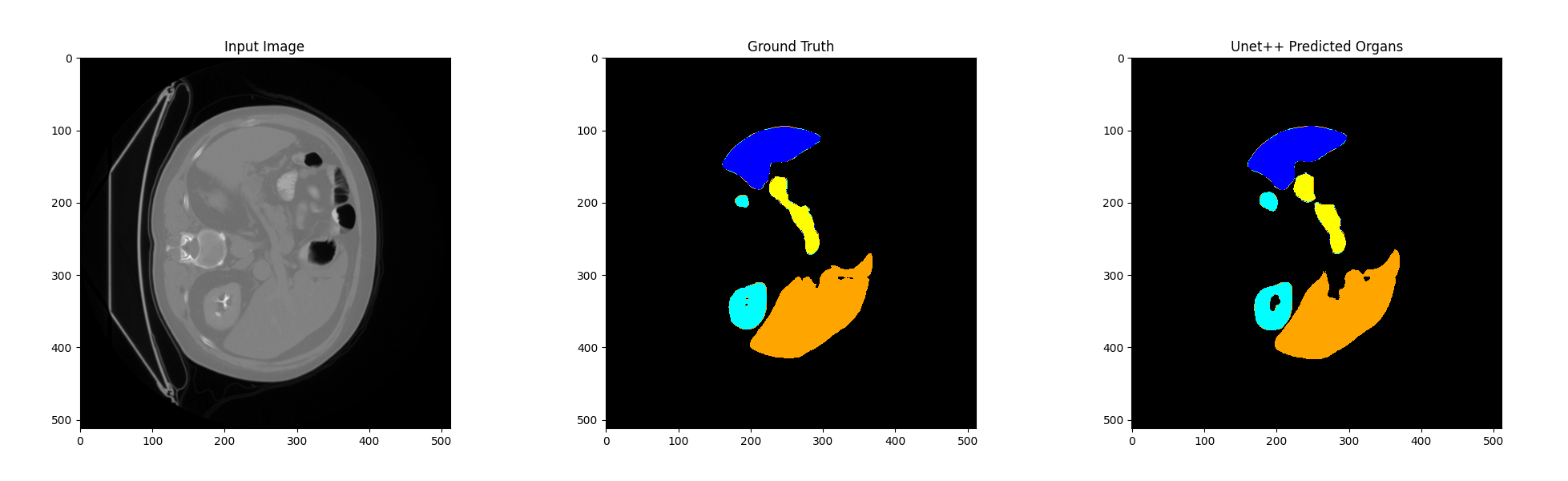
As shown in the Table [???], both models perform consistently well on the liver and the kidney, but for spleen and pancreas, UNet++ demonstrates much better generalization, particularly on the pancreas. TransUNet’s poor performance on these organs in the test set highlights the difficulty of the transformer-based architecture to generalize well on small, variable-shaped organs when trained in a 2D context. The overall results suggest that while TransUNet shows strong performance on the BTCV training set, especially for more straightforward organs like the liver and spleen, it struggles to generalize to unseen data from the AbdomenCT-1K test set. The model’s Dice scores drop notably for challenging organs such as the spleen and pancreas, indicating potential overfitting to the BTCV training data. In contrast, UNet++ demonstrates better generalization across the test set. Compared with the results of training the model directly in 3D, this 2D slicing method reduces the difficulty of model training to some extent, but the results are far from 3D.
3.2. Discussion
3.2.1. 3D Discussion
The nnU-Net, which relies on convolutional blocks, excels at capturing local spatial context and is widely recognized and used globally. CSA-nnU-Net builds upon nnU-Net by adding an attention block without altering other aspects of the model. In contrast, nnFormer utilizes volume attention, allowing it to effectively learn long-range relationships and integrate contextual information, which is crucial for accurately segmenting large 3D CT images, particularly within the bottleneck block. Following the guidelines provided in [10], we computed metrics including Dice, Accuracy, AUC and Volumetric similarity. The four metrics selected show a non-strong correlation to reflect the evaluation of the model prediction results from different aspects [11]. Additionally, we generated violin plots to illustrate the distribution across the dataset. Thus, more specific explanations can be given.
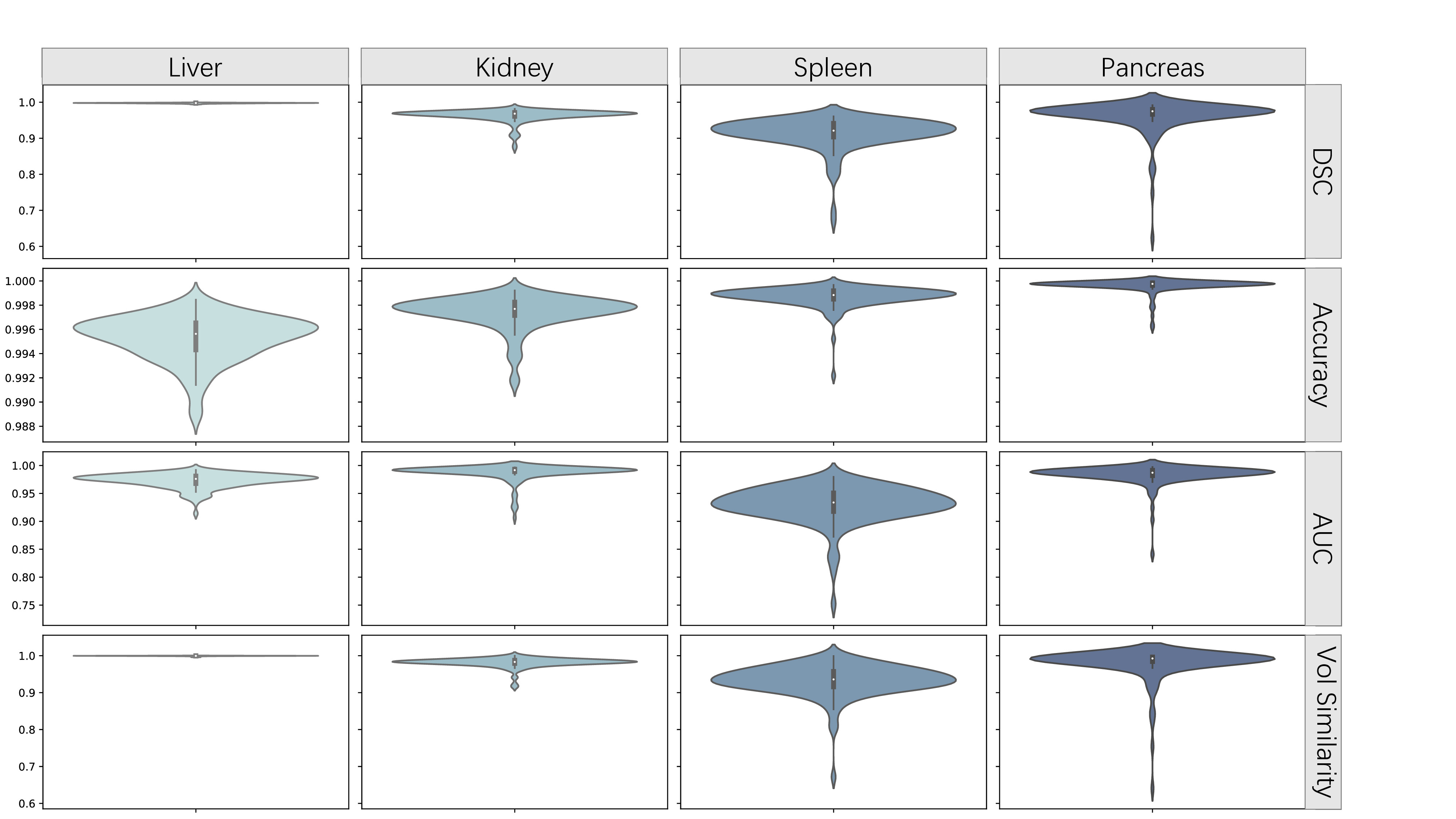
Overall, the different metrics for the three models exhibit shapes that approximate a normal distribution. In the segmentation of the first three classes, the predictions made by nnFormer (Figure 6) across the four metrics are closer to 1 and more concentrated, particularly with no outlier results. This is crucial in medical image segmentation, as it indicates that the model is less likely to produce anomalous results when dealing with CT scans captured under different conditions, thereby avoiding potentially severe consequences. This suggests that nnFormer demonstrates better robustness during transfer learning. In the segmentation of the pancreas, the metrics reveal that the UNet-based models outperform nnFormer. Although less pronounced, nnU-Net (Figure [???]) slightly outperforms CSA-nnU-Net (Figure [???]). The nnFormer, compared to other models, exhibits more outliers in the pancreas segmentation, with distributions ranging from 0 to near-normal at the tails. This indicates that while the model has learned the main features of this class, it lacks generalization and detailed understanding, resulting in poorer performance in terms of transferability and robustness. As for CSA-nnU-Net, it did not demonstrate a clear advantage in any class. Although it performed the worst on the training set, it showed results comparable to the other two models on the test set. While this indicates its relatively strong robustness, the final results were still unsatisfactory. Therefore, under the current experimental conditions, CSA-nnU-Net was unable to fully realize its potential. However, it is important to note that all models predict some non-fully-connected organ segments, which is an issue that can potentially be addressed using post-processing methods. Considering instances where model performance was suboptimal, we found that model robustness is closely linked to the type of basic blocks used. Since convolutions are adept at capturing local features, UNet-based models in our experiments tended to mistakenly classify the spleen as the liver due to local similarities, leading to reduced robustness. However, for smaller organs like the pancreas, the ability of convolutions to quickly capture spatial detail gives them an advantage in transfer learning scenarios. In contrast, nnFormer is able to avoid the aforementioned errors. However, during generalization, nnFormer exhibited an unusual increase in the Dice score for the pancreas, indicating some overfitting given the limited training set and the small size of the organ. As a result, its robustness across different organs is uneven. We believe that for nnFormer, increasing the diversity of the training set should be considered to compensate for the lack of generalization to the pancreas caused by overfitting. As for CSA-nnU-Net, the tolerance for early stopping should be increased; otherwise, it may combine the weaknesses of both convolutional and transformer models rather than their strengths.
3.2.2. 2D Discussion
When medical imaging data is reduced from 3D to 2D formats for analysis, crucial spatial cues between the layers are inevitably diminished. This dimensionality reduction can hinder the model's ability to accurately discern and delineate multi-slice structures like the liver or pancreas. Utilizing 3D Convolutional Neural Networks for medical image analysis allows for a comprehensive exploitation of volumetric data, thereby improving segmentation precision. In contrast, 2D models, which lack depth perception, are disadvantaged when it comes to discerning the intricate spatial interdependencies inherent in 3D datasets. This limitation can hinder the model's efficacy in learning the nuances of complex 3D anatomical configurations or those exhibiting substantial variation across different slices. During the training phase, the granularity and visibility of the anatomical features can fluctuate significantly from one slice to another within the same 3D volume. As 2D slice-based models process each slice in isolation, they risk overlooking the subtleties of the target anatomy when it is either diminutive or poorly defined in certain slices. This variability can skew the training process, potentially leading to suboptimal model performance, especially when the model is exposed to a preponderance of slices where the target structure is less discernible. Anatomical entities like the pancreas might be partially visible or entirely absent in some slices, which can exacerbate the training data imbalance. Such imbalance can skew the model's learning, causing it to overfit on more prevalent classes while struggling with less frequent or less conspicuous samples, such as the pancreas in certain slices. In conclusion, the transition from 3D to 2D for medical image training often compromises the segmentation accuracy due to the absence of spatial context, the fragmentation of anatomical integrity, and the incapacity to leverage depth information effectively. These limitations can result in less precise segmentation outcomes, particularly for structures with intricate or irregular anatomical forms, when employing 2D slice-based models.
4. Conclusion
Our study highlights that 3D models consistently outperform 2D models in capturing spatial relationships and delivering more accurate segmentation results. The 3D models generally outperform their performance on the training set when tested on the test set, a feature that is not as evident in the 2D models. Convolutional models excel in segmenting small and complex structures due to their strong local feature extraction capabilities, while transformer-based models demonstrate advantages in handling larger and more cohesive anatomical regions. The choice of model architecture and dimensional approach should therefore be carefully considered based on the specific requirements of the medical imaging task. Moreover, under the established training conditions, the attention mechanism did not improve the model's performance. Careful consideration of the features of the training and test sets, as well as the training conditions, should be taken before introducing the attention mechanism.
5. Future Work
Given that many factors influence robustness, our incomplete experimental results have certain limitations. In future work, we suggest the following:
- [1.] Increase the diversity of test datasets: This will allow us to evaluate how the characteristics of the test datasets (and their differences from the training sets) affect model robustness. [2.] Expand the number of experimental models: Introduce additional models that combine convolutional UNet structures with ViT structures to further investigate the impact of basic blocks on robustness. [3.] Test model robustness under different training conditions: Assess robustness with varying training durations, epochs, etc., and conduct cross-comparisons with the different models mentioned in the previous point.
Acknowledgement Chuhan Liu, Zicheng Lin, Yang Zhao, Jingkun Shi, Ruoyi Li contributed equally to this work and should be considered co-first authors. Thanks to Professor Jens Rittscher, Institute of Biomedical Engineering, University of Oxford, for his guidance.
References
[1]. Clarivate. Web of science platform. https://clarivate. com/products/scientific-and-academic-research/ research-discovery-and-workflow-solutions/webofscience-platform/ #resources. Accessed: 2024-08-08.
[2]. Synapse. Multi-atlas labeling beyond the cranial vault - workshop and challenge, 2015.
[3]. Jun Ma, Yao Zhang, Song Gu, Cheng Zhu, Cheng Ge, Yichi Zhang, Xingle An, Congcong Wang, Qiyuan Wang, Xin Liu, Shucheng Cao, Qi Zhang, Shangqing Liu, Yunpeng Wang, Yuhui Li, Jian He, and Xiaoping Yang. Abdomenct-1k: Is abdominal organ segmentation a solved problem? IEEE Transactions on Pattern Analysis and Machine Intelligence, 44(10):6695–6714, 2022.
[4]. Hong-Yu Zhou, Jiansen Guo, Yinghao Zhang, Xiaoguang Han, Lequan Yu, Liansheng Wang, and Yizhou Yu. nnformer: Volumetric medical image segmentation via a 3d transformer. IEEE Transactions on Image Processing, 32:4036–4045, 2023.
[5]. Fabian Isensee, Paul F Jaeger, Simon AA Kohl, Jens Petersen, and Klaus H Maier-Hein. nnu-net: a self-configuring method for deep learning-based biomedical image segmentation. Nature methods, 18(2):203–211, 2021.
[6]. Niccolo` McConnell, Nchongmaje Ndipenoch, Yu Cao, Alina Miron, and Yongmin Li. Exploring advanced architectural variations of nnunet. Neurocomputing, 560:126837, 2023.
[7]. Jieneng Chen, Jieru Mei, Xianhang Li, Yongyi Lu, Qihang Yu, Qingyue Wei, Xiangde Luo, Yutong Xie, Ehsan Adeli, Yan Wang, et al. Transunet: Rethinking the u-net architecture design for medical image segmentation through the lens of transformers. Medical Image Analysis, page 103280, 2024.
[8]. Zongwei Zhou, Md Mahfuzur Rahman Siddiquee, Nima Tajbakhsh, and Jianming Liang. Unet++: Redesigning skip connections to exploit multiscale features in image segmentation. IEEE Transactions on Medical Imaging, 2019.
[9]. Adam K. Wolf. 3d nii visualizer. https://github.com/adamkwolf/ 3d-nii-visualizer, 2024. Accessed: 2024-09-04.
[10]. Dominik Mu ̈ller, In ̃aki Soto-Rey, and Frank Kramer. Towards a guideline for evaluation metrics in medical image segmentation. BMC Research Notes, 15(1):210, 2022.
[11]. Abdel Aziz Taha and Allan Hanbury. Metrics for evaluating 3d medical image segmentation: analysis, selection, and tool. BMC medical imaging, 15:1–28, 2015.
Cite this article
Liu,C.;Lin,Z.;Zhao,Y.;Shi,J.;Li,R. (2025). The Robustness Evaluation of Advanced Models in CT Image Segmentation: From Multi-organ to Multi-organ. Applied and Computational Engineering,132,306-313.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 2nd International Conference on Machine Learning and Automation
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Clarivate. Web of science platform. https://clarivate. com/products/scientific-and-academic-research/ research-discovery-and-workflow-solutions/webofscience-platform/ #resources. Accessed: 2024-08-08.
[2]. Synapse. Multi-atlas labeling beyond the cranial vault - workshop and challenge, 2015.
[3]. Jun Ma, Yao Zhang, Song Gu, Cheng Zhu, Cheng Ge, Yichi Zhang, Xingle An, Congcong Wang, Qiyuan Wang, Xin Liu, Shucheng Cao, Qi Zhang, Shangqing Liu, Yunpeng Wang, Yuhui Li, Jian He, and Xiaoping Yang. Abdomenct-1k: Is abdominal organ segmentation a solved problem? IEEE Transactions on Pattern Analysis and Machine Intelligence, 44(10):6695–6714, 2022.
[4]. Hong-Yu Zhou, Jiansen Guo, Yinghao Zhang, Xiaoguang Han, Lequan Yu, Liansheng Wang, and Yizhou Yu. nnformer: Volumetric medical image segmentation via a 3d transformer. IEEE Transactions on Image Processing, 32:4036–4045, 2023.
[5]. Fabian Isensee, Paul F Jaeger, Simon AA Kohl, Jens Petersen, and Klaus H Maier-Hein. nnu-net: a self-configuring method for deep learning-based biomedical image segmentation. Nature methods, 18(2):203–211, 2021.
[6]. Niccolo` McConnell, Nchongmaje Ndipenoch, Yu Cao, Alina Miron, and Yongmin Li. Exploring advanced architectural variations of nnunet. Neurocomputing, 560:126837, 2023.
[7]. Jieneng Chen, Jieru Mei, Xianhang Li, Yongyi Lu, Qihang Yu, Qingyue Wei, Xiangde Luo, Yutong Xie, Ehsan Adeli, Yan Wang, et al. Transunet: Rethinking the u-net architecture design for medical image segmentation through the lens of transformers. Medical Image Analysis, page 103280, 2024.
[8]. Zongwei Zhou, Md Mahfuzur Rahman Siddiquee, Nima Tajbakhsh, and Jianming Liang. Unet++: Redesigning skip connections to exploit multiscale features in image segmentation. IEEE Transactions on Medical Imaging, 2019.
[9]. Adam K. Wolf. 3d nii visualizer. https://github.com/adamkwolf/ 3d-nii-visualizer, 2024. Accessed: 2024-09-04.
[10]. Dominik Mu ̈ller, In ̃aki Soto-Rey, and Frank Kramer. Towards a guideline for evaluation metrics in medical image segmentation. BMC Research Notes, 15(1):210, 2022.
[11]. Abdel Aziz Taha and Allan Hanbury. Metrics for evaluating 3d medical image segmentation: analysis, selection, and tool. BMC medical imaging, 15:1–28, 2015.









