1. Introduction
Medical named entity recognition aims to identify all named entities of a specific clinically relevant type in a given unstructured clinical report. Using medical-named entity recognition, one can further analyze, aggregate, and mine valid information from electronic medical records (EHRs) [1]. Nowadays, EHRs have become important documents for doctors to diagnose and record patients' conditions and medical history, and by using automatic entity recognition in EHRs, the time taken by doctors to collate information can be reduced by more than 80%.
Previous research has developed many models for medical named entity recognition, including BiLSTM-CRF models, BERT-CRF, and Bio BERT. The current state-of-the-art NER system uses a pre-trained neural architecture based on a language modelling task [2-4]. By using language model pre-training, a significant improvement in the accuracy of named entity recognition has been achieved.
However, previous methods do not use dynamic fusion weights, which makes the complementary nature of features in CNN, BiLSTM and transformer not fully demonstrated, and the potential of named entity recognition methods in the medical field is still in the exploratory stage. Therefore, in this paper, we propose a novel Bidirectional Gated Pyramid Network (BGPN), which dynamically assigns weights to the CNN, BiLSTM, and Transformer layers, fuses the features of all three, and combines them with a CRF to output the optimal prediction sequence of BIO label. The model innovatively combines character-level features, features between upper and lower sentences, and multi-level features of full-text information, which provides a new idea for research in this field.
2. Related work
Early models used for named entity recognition are the Hidden Markov Model (HMM), CRF, etc [5-6]. Later, CNN-based models were proposed to solve the sequence labelling problem, which inspired many subsequent models to combine CNN and CRF, which became early benchmarks for deep learning in NER tasks. Then, recurrent neural network-based models that were more suitable for processing sequences appeared. Huang et al. proposed for the first time to combine a BiLSTM with a CRF layer for sequence labelling tasks to solve the problem of insufficient modelling of subsequent word dependencies by traditional unidirectional LSTMs [7].
The most popular named entity recognition systems currently use neural network architectures pre-trained on language modelling tasks. Such example is Bio BERT, a variant of BERT pre-trained on the PubMed corpus, which significantly improves the accuracy of recognizing entities such as diseases, genes, etc [8]. In addition to this, there are models that incorporate graph convolutional networks, such as GCN-BiLSTM [9].
However, existing methods do not adequately incorporate the dynamic fusion of multi-level features, such as dynamic fusion relationships between character layers, inter-sentence context, and full-text information. Therefore, the model proposed in this paper uses a gating mechanism to dynamically weight the CNN layer, BiLSTM layer, and Transformer layer to avoid manually designing the fusion rules and calculating the label transfer probability from the training data to constrain the unreasonable transfer.
3. Methods
3.1. Data preprocessing
The BC5CDR dataset contains 1,500 abstracts from PubMed, which is equally divided into a training set, a validation set, and a test set (500 abstracts each) [10]. Figure 1 illustrates the data preprocessing process for this dataset. Firstly, the label format was converted into the standard BIO format, which contains five labels respectively B-Disease I-Disease B-Chemica I-Chemical O. Then the text sentence sequences and the corresponding label sequences were obtained separately. The text sentence sequences are then subjected to character embedding (20-dimensional vectors for each character) and word embedding (50-dimensional vectors for each word), and finally, they are fed into the CNN layer and BiLSTM layer, respectively.
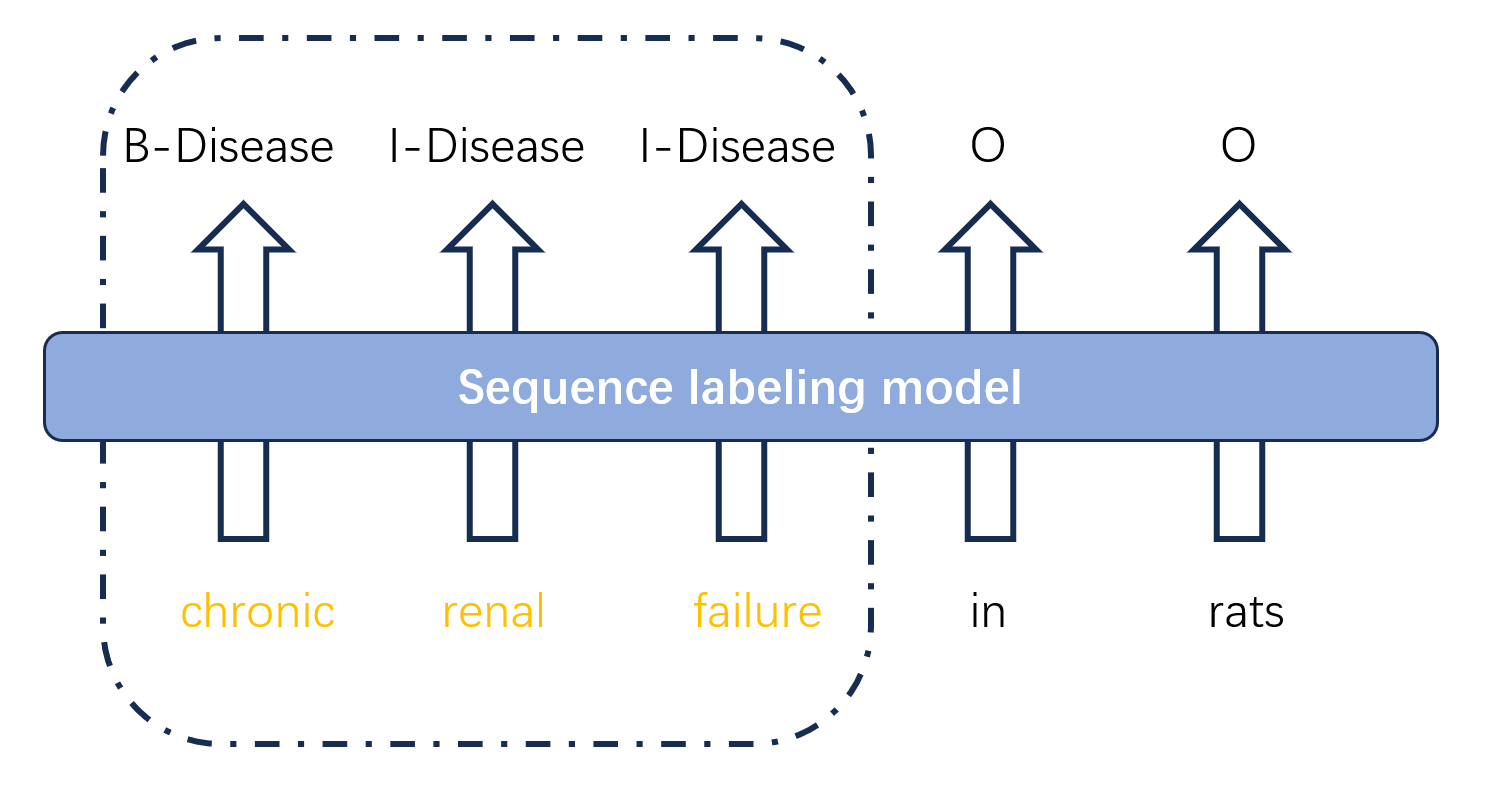
Figure 1: Converting data to BIO label format
3.2. Model building
Figure 2 illustrates the overall architecture of the BGPN. The network consists of three parts, specifically, the bottom layer of the network is a CNN layer for extracting character-level features, the middle layer is a BiLSTM layer capturing local inter-contextual sentence connections, and the top layer of the network is the encoder portion of a transformer for capturing the global contextual information of the text. Subsequently, we propose the Gated Fusion Layer, where the output feature vectors of each layer are given a corresponding weight, and further weighted fusion in that layer yields a fused feature. The features are further fed into the Fully Connected Layer and the SoftMax layer to get predicted scores for five categories. Finally, these scores are fed into the CRF layer to generate the optimal prediction sequence.
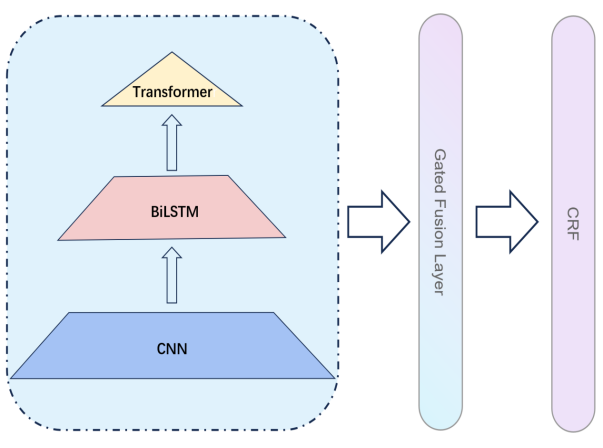
Figure 2: Overall architecture of bidirectional gated pyramid network
3.2.1. CNN
Convolutional Neural Networks (CNNs) have a significant advantage in character-level semantic modelling due to their local feature extraction capability, which can identify abbreviations of medical terms, e.g., ASA can be identified as an abbreviation of Aspirin [11]. The CNN layer of this model uses multi-scale convolutional kernels of sizes 3, 5, and 7 to build a hierarchical perception mechanism, with the convolution of size 3 focusing on the combinatorial patterns of locally adjacent characters (e.g., "A-S" and "S-A" binary connections in "ASA"), the convolutional kernel of size 5 capturing the associative features of medium-spanning characters (e.g., prefix/suffix structures), and the convolution of size 7 modelling long-range character dependencies (e.g., cross-interval chemical dependencies).character dependencies (e.g., chemical formula fragments across intervals). The input is a 20-dimensional character embedding vector, which is subjected to parallel feature extraction through these three sets of independent convolutional layers, and then the most salient features are retained by Adaptive Max Pooling and finally projected to a 128-dimensional feature space after two linear layers.
3.2.2. BiLSTM
In sequence annotation tasks, we can obtain past and future input features for a given point in time, so we can use a BiLSTM network [12]. The BiLSTM layer splices the output vectors obtained from the CNN layer with the word embedding vectors to obtain a 178-dimensional vector as input, with the hidden unit set to 64, which is passed through a forward LSTM layer and an inverse LSTM layer to learn the information from the previous tokens as well as the subsequent tokens, and finally, a 128-dimensional vector containing inter-sentence contextual information features is obtained [13].
3.2.3. Transformer
Attention mechanisms have become an integral part of compelling sequence modelling and transformation models for a variety of tasks, allowing modelling while ignoring dependency distances in input or output sequences [15]. Considering the need to capture global sequence dependencies, the highest layer of our model is the encoder part of the Transformer. The Transformer layer takes the output vector of the BiLSTM layer as input, configured with a 4-head self-attention mechanism to achieve multidimensional feature interactions, and the feedforward neural network is extended to 256 dimensions to enhance nonlinear representations, with a 0.3 dropout ratio for regularization control, and finally outputs 128-dimensional global context-aware vectors.
3.2.4. Gated fusion layer
Previous feature fusion strategies relying on manual design (e.g., simple splicing or static weighting) did not consider the dynamic interaction of features at different levels, and the weights corresponding to features could not be dynamically updated following the training of the model. For this reason, this paper introduces a gated fusion mechanism to dynamically assign weights for feature representations of character CNN, BiLSTM and Transformer. Specifically, to obtain the outputs of each layer of the pyramid network, the feature \( C∈{R^{b×n×{d_{c}}}} \) for the CNN layer, the feature \( L∈{R^{b×n×{d_{l}}}} \) for the BiLSTM layer, and the feature \( T∈{R^{b×n×{d_{t}}}} \) for the Transformer layer (where \( b \) is the batch size, and \( n \) is the length of the sequence). firstly, after splicing the three features together, we obtain the feature \( F=[C;L;T]∈{R^{b×n×({d_{c}}+{d_{l}}+{d_{t}})}} \) , followed by generating the dynamic weights corresponding to the CNN layer, BiLSTM layer, and Transformer layer by a linear transform: \( G=Softmax({W_{g}}F+{b_{g}})∈{R^{b×n×3}} \) . The fusion feature \( H \) is generated by weighted summation: \( H=\sum _{k=1}^{3} {G_{k}}⊙{F_{k}}∈{R^{b×n×{d_{h}}}} \) , where \( {F_{k}} \) corresponds to \( C \) , \( L \) , and \( T \) , respectively, and \( ⊙ \) denotes element-by-element multiplication. The predicted scores for five categories are finally obtained through the fully connected layer. Compared to the fixed weight assignment, this mechanism allows the model to dynamically adjust the contribution of each module according to the context, e.g., to enhance the role of Character CNNs when recognizing complex medical terms and to focus on Transformer features when dealing with long-distance dependencies.
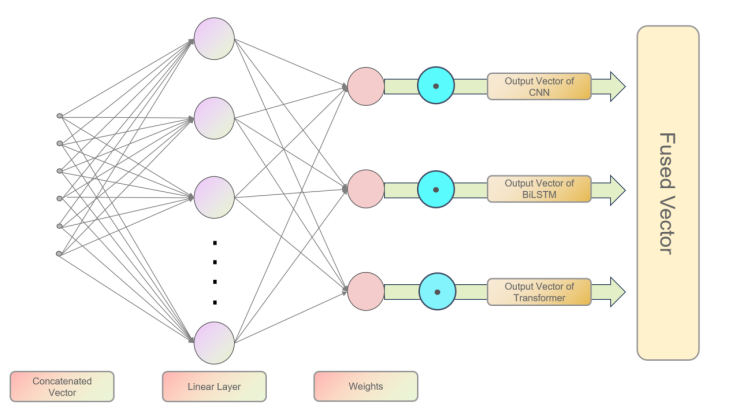
Figure 3: The details of our proposed gated fusion layer
3.2.5. CRF
To model the transfer dependencies between labels and constrain the global plausibility of sequence prediction, this model uses conditional random as the final decoder [16]. The predicted scores for five categories output from the gated fusion layer are fed into the CRF layer, and the optimal prediction sequence of BIO labels is finally obtained by dynamic planning with the Viterbi algorithm [17].
4. Experiment and analysis
This experiment evaluates the model performance based on the BC5CDR dataset, with the training configuration containing the AdamW optimizer (initial learning rate of 2e-3, weight decay set to 0.05) and the learning rate dynamically adjusted during training.
Figure 4 illustrates the change in LOSS on both the training and validation sets. During the first fifteen rounds of training, the overall LOSS on both the training and validation sets decreases, and when the training rounds reach the tenth round, the LOSS on the validation set decreases insignificantly, and the number of training rounds for the model is chosen to be 15 in order to reduce the overfitting. It is interesting to note that the validation loss is always higher than the training loss; especially after Epoch 3, the difference gradually widens. This may be due to two reasons. One is the mismatch between model complexity and data size; the training set of BC5CDR has only 500 PubMed documents, and high-capacity models are prone to memory noise on small data. The second is that the data preprocessing only fills the data and does not introduce medical text-specific enhancement operations [18].
Table 1 shows the results of testing the performance of the present model on the test set. The present model is significantly ahead in recognizing chemical entities, indicating that the model is reliable in predicting the ortho-class of chemical entities, but the recall is low at only 0.70, with a high number of misses, probably due to the weak recognition of chemical abbreviations as well as variants. It should be noted that the precision and recall of disease are both lower than chemical at 0.71 and 0.69 respectively, indicating that the model has double difficulties in describing the disease, and possible reasons for this include the high number of nested entities in disease and the fact that the recognition of disease is very dependent on the global context.
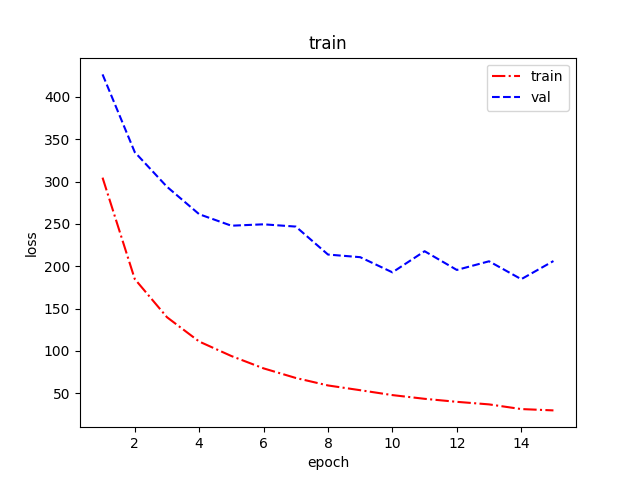
Figure 4: Loss per epoch during training
Table 1: The performance of our method evaluated on test dataset
Precision | Recall | F1-score | |
Chemical | 0.91 | 0.70 | 0.79 |
Disease | 0.71 | 0.69 | 0.70 |
Micro avg | 0.81 | 0.69 | 0.75 |
Macro avg | 0.81 | 0.69 | 0.74 |
Weighted avg | 0.82 | 0.69 | 0.75 |
5. Conclusion
This paper proposes a novel Bidirectional Gated Pyramid Network (BGPN), which innovatively uses a gated fusion mechanism to dynamically fuse character-level, sentence-level, and long-distance context features at multiple levels, and combines with a conditional random field to constrain unreasonable label sequences by statistically shifting the probability of label transfer, and finally obtain the optimal label sequence. The proposed algorithm provides a useful idea for the research in this area, as it finally obtains the optimal tag sequence. However, due to the small dataset and high complexity of the model, the model did not achieve very satisfactory results on the test set. Future improvements include data enhancement, introduction of medical synonym replacement, and adversarial sample generation. Model optimization can also be performed, such as changing the random initialization of the word embedding part to word embedding using a pre-trained model.
References
[1]. Kundeti, S. R., Vijayananda, J., Mujjiga, S., & Kalyan, M. (2016, December). Clinical named entity recognition: Challenges and opportunities. In 2016 IEEE International Conference on Big Data (Big Data)(pp. 1937-1945). IEEE.
[2]. Huang, Z., Xu, W., & Yu, K. (2015). Bidirectional LSTM-CRF models for sequence tagging. arXiv preprint arXiv:1508.01991.
[3]. Souza, F., Nogueira, R., & Lotufo, R. (2019). Portuguese named entity recognition using BERT-CRF. arXiv preprint arXiv:1909.10649.
[4]. Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H., & Kang, J. (2020). BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics, 36(4), 1234-1240.
[5]. Morwal, S., Jahan, N., & Chopra, D. (2012). Named entity recognition using hidden Markov model (HMM). International Journal on Natural Language Computing (IJNLC) Vol, 1.
[6]. Lafferty, J., McCallum, A., & Pereira, F. (2001, June). Conditional random fields: Probabilistic models for segmenting and labeling sequence data. In Icml (Vol. 1, No. 2, p. 3).
[7]. Huang, Z., Xu, W., & Yu, K. (2015). Bidirectional LSTM-CRF models for sequence tagging. arXiv preprint arXiv:1508.01991.
[8]. Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H., & Kang, J. (2020). BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics, 36(4), 1234-1240.
[9]. Sui, D., Chen, Y., Liu, K., Zhao, J., & Liu, S. (2019, November). Leverage lexical knowledge for Chinese named entity recognition via collaborative graph network. In Proceedings of the 2019 conference on empirical methods in natural language processing and the 9th international joint conference on natural language processing (EMNLP-IJCNLP) (pp. 3830-3840).
[10]. Li, J., Sun, Y., Johnson, R. J., Sciaky, D., Wei, C. H., Leaman, R., ... & Lu, Z. (2016). BioCreative V CDR task corpus: a resource for chemical disease relation extraction. Database, 2016.
[11]. Zhang, X., Zhao, J., & LeCun, Y. (2015). Character-level convolutional networks for text classification. Advances in neural information processing systems, 28.
[12]. Rhanoui, M., Mikram, M., Yousfi, S., & Barzali, S. (2019). A CNN-BiLSTM model for document-level sentiment analysis. Machine Learning and Knowledge Extraction, 1(3), 832-847.
[13]. Chen, T., Xu, R., He, Y., & Wang, X. (2017). Improving sentiment analysis via sentence type classification using BiLSTM-CRF and CNN. Expert Systems with Applications, 72, 221-230.
[14]. Liu, Y., Liu, N., Wu, Y., Cholakkal, H., Anwer, R. M., Yao, X., & Han, J. (2024). NTRENet++: Unleashing the Power of Non-target Knowledge for Few-shot Semantic Segmentation. IEEE Transactions on Circuits and Systems for Video Technology.
[15]. Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A. N., ... & Polosukhin, I. (2017). Attention is all you need. Advances in neural information processing systems, 30.
[16]. Sutton, C., & McCallum, A. (2012). An introduction to conditional random fields. Foundations and Trends® in Machine Learning, 4(4), 267-373.
[17]. Wallach, H. M. (2004). Conditional random fields: An introduction (Vol. 22, pp. 2-6). Technical reports (CIS).
[18]. Symeonidou, A., Sazonau, V., & Groth, P. (2019). Transfer Learning for Biomedical Named Entity Recognition with BioBERT. In SEMANTiCS (Posters & Demos).
Cite this article
Jiang,S. (2025). Medical Named Entity Recognition Based on Bidirectional Gated Pyramid Network. Applied and Computational Engineering,152,72-77.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 3rd International Conference on Software Engineering and Machine Learning
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Kundeti, S. R., Vijayananda, J., Mujjiga, S., & Kalyan, M. (2016, December). Clinical named entity recognition: Challenges and opportunities. In 2016 IEEE International Conference on Big Data (Big Data)(pp. 1937-1945). IEEE.
[2]. Huang, Z., Xu, W., & Yu, K. (2015). Bidirectional LSTM-CRF models for sequence tagging. arXiv preprint arXiv:1508.01991.
[3]. Souza, F., Nogueira, R., & Lotufo, R. (2019). Portuguese named entity recognition using BERT-CRF. arXiv preprint arXiv:1909.10649.
[4]. Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H., & Kang, J. (2020). BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics, 36(4), 1234-1240.
[5]. Morwal, S., Jahan, N., & Chopra, D. (2012). Named entity recognition using hidden Markov model (HMM). International Journal on Natural Language Computing (IJNLC) Vol, 1.
[6]. Lafferty, J., McCallum, A., & Pereira, F. (2001, June). Conditional random fields: Probabilistic models for segmenting and labeling sequence data. In Icml (Vol. 1, No. 2, p. 3).
[7]. Huang, Z., Xu, W., & Yu, K. (2015). Bidirectional LSTM-CRF models for sequence tagging. arXiv preprint arXiv:1508.01991.
[8]. Lee, J., Yoon, W., Kim, S., Kim, D., Kim, S., So, C. H., & Kang, J. (2020). BioBERT: a pre-trained biomedical language representation model for biomedical text mining. Bioinformatics, 36(4), 1234-1240.
[9]. Sui, D., Chen, Y., Liu, K., Zhao, J., & Liu, S. (2019, November). Leverage lexical knowledge for Chinese named entity recognition via collaborative graph network. In Proceedings of the 2019 conference on empirical methods in natural language processing and the 9th international joint conference on natural language processing (EMNLP-IJCNLP) (pp. 3830-3840).
[10]. Li, J., Sun, Y., Johnson, R. J., Sciaky, D., Wei, C. H., Leaman, R., ... & Lu, Z. (2016). BioCreative V CDR task corpus: a resource for chemical disease relation extraction. Database, 2016.
[11]. Zhang, X., Zhao, J., & LeCun, Y. (2015). Character-level convolutional networks for text classification. Advances in neural information processing systems, 28.
[12]. Rhanoui, M., Mikram, M., Yousfi, S., & Barzali, S. (2019). A CNN-BiLSTM model for document-level sentiment analysis. Machine Learning and Knowledge Extraction, 1(3), 832-847.
[13]. Chen, T., Xu, R., He, Y., & Wang, X. (2017). Improving sentiment analysis via sentence type classification using BiLSTM-CRF and CNN. Expert Systems with Applications, 72, 221-230.
[14]. Liu, Y., Liu, N., Wu, Y., Cholakkal, H., Anwer, R. M., Yao, X., & Han, J. (2024). NTRENet++: Unleashing the Power of Non-target Knowledge for Few-shot Semantic Segmentation. IEEE Transactions on Circuits and Systems for Video Technology.
[15]. Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A. N., ... & Polosukhin, I. (2017). Attention is all you need. Advances in neural information processing systems, 30.
[16]. Sutton, C., & McCallum, A. (2012). An introduction to conditional random fields. Foundations and Trends® in Machine Learning, 4(4), 267-373.
[17]. Wallach, H. M. (2004). Conditional random fields: An introduction (Vol. 22, pp. 2-6). Technical reports (CIS).
[18]. Symeonidou, A., Sazonau, V., & Groth, P. (2019). Transfer Learning for Biomedical Named Entity Recognition with BioBERT. In SEMANTiCS (Posters & Demos).









