1. Introduction
Pneumonia is a lung infection caused by bacteria, viruses, or other microorganisms. It is a common clinical disease that predominantly occurs among the elderly and children. In the field of medical image analysis, traditional diagnostic methods rely heavily on manual feature extraction and expert experience, which are limited by strong subjectivity and high individual variability.
In recent years, with the advancement of deep learning technologies—particularly convolutional neural networks (CNNs), one of the core models in the field—CNNs have gradually become the foundation of image recognition and computer vision. Owing to their three main features—local connectivity, weight sharing, and spatial subsampling—CNNs can effectively process medical images, injecting revolutionary momentum into medical image analysis. Against this background, this study establishes a deep learning environment, builds a neural network model using a pneumonia dataset, and conducts experiments. The results demonstrate that the proposed method is effective in pneumonia detection.
This paper is organized into six sections: Introduction, Literature Review, Methodology, Results, Discussion, and Conclusion. The Introduction highlights the high incidence of pneumonia and the efficiency bottlenecks of traditional diagnosis, while also outlining the application background of CNNs in medical image analysis, thereby laying both the practical and technical foundation for this research. The Literature Review systematically examines the extensive applications of CNNs in the medical field in recent years, clarifying the technical context and frontiers of the study. The Methodology section not only explains the basic principles of CNNs but also provides a detailed account of the AlexNet architecture employed in this research, offering theoretical and technical support for experimental design. The Results section describes the composition of the dataset, key parameter settings, and the accuracy variations of the model under different epoch values. The Discussion section engages in an in-depth analysis of central issues in the research design, including the rationale for selecting this model, the suitability of the dataset, the role of the flatten and dropout layers in the convolutional network, the function of normalization in data preprocessing, and the observed trends of experimental performance across different epochs. Finally, the Conclusion summarizes the study, objectively points out its limitations, and proposes directions for future optimization.
2. Literature review
Convolutional neural networks (CNNs) play a key role in medical image recognition and detection. In recent years, their applications in the medical field have continued to deepen and have been widely implemented in practice. Researchers have developed and applied various CNN architectures for medical image analysis tasks, including basic CNN and improved classification networks, U-shaped and variant segmentation networks, 3D convolutional networks, lightweight networks, and attention-mechanism-based fusion networks.
Basic CNN and improved classification networks. Built upon classical CNN architectures, these models enhance performance in medical image classification tasks by optimizing loss functions, introducing feature enhancement strategies, or employing transfer learning. For example, Kun Hao et al. proposed a novel cost function—weighted Fisher criterion—and applied it to a CNN model for breast cancer detection, achieving an F1-score of 0.920 and demonstrating excellent performance [1]. Later, Xiaomei Shen et al. optimized CNN feature extraction modules for salivary gland tumor ultrasound images, combining gamma transformation with bilateral filtering to enhance texture features, thereby achieving an accuracy of 85.44% and a sensitivity of 86.67%, which reduced unnecessary fine-needle biopsies [2]. Xin Song et al. proposed a hybrid intelligent classification model combining CNN and k-nearest neighbors (KNN). By using transfer learning to optimize parameters, the model achieved an F1-score of 94.61% and an accuracy of 94.73% in sickle cell disease (SCD) image classification, enabling efficient automatic SCD recognition [3].
U-shaped networks and their variants. Centered on the “encoder–decoder with skip connections” architecture, these models improve segmentation accuracy through feature fusion and have become the mainstream method in medical image segmentation. Runhua Shao et al. improved U-Net by introducing an attention-guided connection (AGC) module, which increased the Intersection over Union (IoU) by 5.4% on the PAIP-2019 dataset. With multi-scale dilated convolution for cross-level feature fusion, the model achieved a Dice coefficient of 0.863, significantly enhancing tumor region segmentation [4]. Panpan Liu et al. proposed a novel asymmetric U-shaped CNN, ASUNet, which achieved Dice scores of 77.08%, 90.83%, and 83.41% for enhancing tumor (ET), whole tumor (WT), and tumor core (TC) regions, respectively, on the BraTS 2020 dataset, with a Hausdorff distance as low as 4.92 mm [5]. Xiaoqin Wu et al. combined U-Net with the watershed algorithm for brain tumor segmentation, achieving an accuracy of 0.896 and specificity of 0.9888, effectively addressing the edge-blurring problem of U-Net [6].
3D convolutional networks. By applying three-dimensional convolution operations, these models capture volumetric spatial information and better model lesion morphology. Qiwei Cao et al. proposed a method based on 3D multi-pooling CNN, which integrates multi-scale input and downsampling strategies with conditional random fields (CRF) for boundary optimization. On 100 multimodal MRI brain tumor segmentation cases, the model achieved a Dice coefficient of 91.64%, effectively accommodating tumor size variations across image slices [7]. Lijun Xu et al. developed a 3D-Ghost convolution module based on 3D U-Net, which reduces redundant features via linear operations, and incorporated a 3D coordinate attention module to enhance spatial awareness. On the BraTS 2018 dataset, Dice scores for WT, TC, and ET regions reached 0.8632, 0.8473, and 0.8036, respectively, with a significant reduction in model parameters [8]. Fei Chen et al. built upon the 3D V-Net framework by introducing a CoordConv layer to fuse image and coordinate information, combined with a semi-supervised Mean Teachers strategy to leverage unlabeled data. In pancreatic tumor segmentation, the model achieved a tumor Dice score of 0.722 ± 0.290 and a Kappa coefficient of 0.746, enabling precise segmentation of pancreatic head–neck, body–tail, and tumor regions [9].
Lightweight networks. These models optimize convolution operations and reduce parameters, achieving higher computational efficiency without sacrificing accuracy. Gang Pei et al. improved the U-shaped architecture by replacing bottleneck convolutions with linear mapping and attention mechanisms, introducing a lightweight multilayer perceptron (Tok-MLP) for positional information learning, and enlarging the receptive field using dilated convolution. Additionally, gated attention was incorporated into skip connections to enhance feature propagation. The proposed model outperformed mainstream algorithms on the BUSI and ISIC2018 datasets in Dice scores, while requiring only 0.74M parameters and 0.40G FLOPs, achieving a balance between segmentation performance and computational efficiency [10].
Attention-mechanism fusion networks. By integrating attention modules, these networks improve focus on key features and effectively suppress noise interference. Taojie Zhang et al. proposed ECENet, which prunes the CHRNet architecture to optimize network structure, incorporates the SKSAM attention module (combining channel and spatial attention), and employs a context-aware fusion block (CoFusion) for multi-scale output integration. On blurry skeletal images, ECENet achieved an ODS of 0.816 and OIS of 0.823, with peak signal-to-noise ratio (PSNR) improved by 16.8% and structural similarity index (SSIM) improved by 37.6% [11].
3. Methodology
3.1. Convolutional Neural Networks
Convolutional Neural Networks (CNNs) are a class of deep learning models specifically designed for structured data processing and are widely applied in image recognition, object detection, and related domains. CNNs consist of multiple hierarchical layers, the most common of which include the input layer, convolutional layers, pooling layers, fully connected layers, and the output layer.
The convolutional layer extracts local features from the input data through convolution operations. Each convolutional layer contains multiple convolution kernels (filters), each of which can be regarded as a linear function composed of weights and bias. The basic principle of convolution layers is to generate feature maps of the input data via kernel-based convolution operations. The computation can be generally expressed as:
where
The pooling layer performs downsampling on the feature maps produced by convolution layers, thereby reducing the number of parameters and compressing computations. This helps mitigate overfitting. Common pooling strategies include average pooling and max pooling. Average pooling computes the mean value within a receptive region, whereas max pooling selects the maximum value. In this study, max pooling layers are employed.
The fully connected layer connects each node to all nodes in the preceding layer, integrating the extracted features into a final classification decision. In CNNs, fully connected layers transform extracted image features into a one-dimensional vector. The computation of a fully connected layer can be expressed as:
3.2. Alexnet
AlexNet was proposed in 2012 by Krizhevsky and colleagues at the University of Toronto [12]. The network is composed of five convolutional layers, three max-pooling layers, and three fully connected layers.
|
Layer |
Hyperparameters |
|
Convolutional |
Filter=(11,11), Stride=4, Padding=0 |
|
Max Pooling |
Kernel size=(3,3), Stride=2 |
|
Convolutional |
Filter=(5,5), Stride=1, Padding=2 |
|
Max Pooling |
Kernel size=(3, 3), Stride=2 |
|
Convolutional |
Filter=(3, 3), Stride=1, Padding=1 |
|
Convolutional |
Filter=(3, 3), Stride=1, Padding=1 |
|
Convolutional |
Filter=(3, 3), Stride=1, Padding=1 |
|
Max Pooling |
Kernel size=(3, 3), Stride=2 |
|
Flatten Layer |
/ |
|
Fully Connected |
(9216,4096) |
|
Dropout |
P=0.5 |
|
Fully Connected |
(4096,4096) |
|
Dropout |
P=0.5 |
|
Fully Connected |
(4096,2) |
4. Results
The dataset used in this study was collected from chest X-ray images, consisting primarily of radiographs from both healthy individuals and patients diagnosed with pneumonia. All images were grayscale. In total, 5,840 chest X-ray images were included. The dataset was divided into two folders, namely train and test, containing 5,216 and 624 images, respectively. Each folder was further divided into two subfolders labeled NORMAL and PNEUMONIA. Specifically, the train folder contained 3,875 images under PNEUMONIA and 1,341 images under NORMAL, while the test folder contained 390 PNEUMONIA images and 234 NORMAL images. Figure 1 illustrates several representative samples from the dataset, where NORMAL indicates a healthy condition and PNEUMONIA indicates the presence of pneumonia.
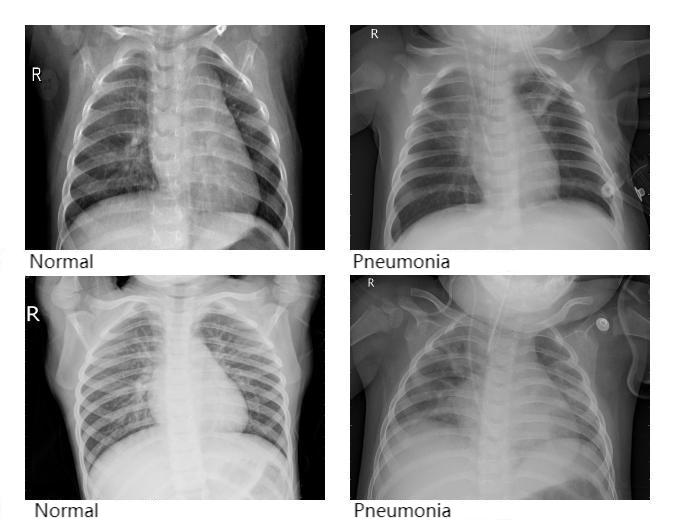
Prior to training, all images were preprocessed as follows: image sizes were resized to 224 × 224 pixels and normalized with mean = 0.5 and standard deviation = 0.5.
For model training, the Adam optimizer was adopted, and cross-entropy loss was chosen as the loss function. The batch size was set to 32 for both the training and validation sets. The learning rate was fixed at 0.0001. To evaluate the impact of training duration, the model was trained under different epoch settings, and performance was compared accordingly. The classification accuracy under different epochs is summarized in Table 2.
|
epoch |
Accuracy on Test Set |
|
1 |
0.7548 |
|
2 |
0.7916 |
|
3 |
0.7147 |
|
4 |
0.8108 |
|
5 |
0.7612 |
|
7 |
0.7371 |
|
10 |
0.7644 |
As shown in Table 2, the model achieved the highest accuracy at epoch = 4, reaching 0.8108.
5. Discussion
With the advancement of deep learning, Convolutional Neural Networks (CNNs) have demonstrated remarkable superiority. From LeNet-5, which leveraged local receptive fields and weight sharing for efficient handwritten digit recognition, to AlexNet, which with its deeper architecture outperformed traditional methods in the ImageNet competition, proving its strong feature extraction capability, and further to the continuous innovations of VGG and ResNet, CNNs have consistently pushed the frontier of image analysis. Compared with traditional image processing methods that rely on handcrafted features, CNNs benefit from large-scale data-driven training, enabling them to dynamically adjust feature extraction strategies and adapt naturally to diverse tasks [13]. For this reason, this study employed a CNN-based model.
Pneumonia is common among individuals with compromised immune systems, often presenting with symptoms such as high fever and cough. Without timely intervention, the condition can progress to severe pneumonia, leading to acute respiratory distress or even shock, thereby posing a direct threat to patient survival [14]. If CNNs can achieve high-accuracy early prediction of pneumonia, they can provide a critical window for earlier intervention and treatment. This justifies the focus of this study on pneumonia-related image analysis.
In the experimental model, flatten and dropout layers were incorporated. The flatten layer converts multi-dimensional feature maps into one-dimensional vectors, serving as a bridge that allows the features extracted by convolutional and pooling layers to be fed into the fully connected layers. The dropout layer, on the other hand, randomly discards units during training, effectively mitigating the risk of overfitting.
Additionally, normalization preprocessing was applied to the dataset prior to training. On the one hand, normalization compressed the dynamic range of pixel values, balancing the differences between bright and dark regions of the image. This prevented overexposure or underexposure from dominating the feature learning process and allowed the model to focus more on structural characteristics of the image. On the other hand, normalization scaled raw data (e.g., pixel intensities and feature values) into a specific range (such as 0–1 or –1–1), enabling the model to more precisely capture subtle feature differences within this sensitive interval, thereby improving classification accuracy.
Furthermore, training the model under different epoch settings revealed a fluctuating trend in accuracy on the test set (see Table 2). Accuracy peaked at epoch 4 with a value of 0.8108, after which it declined from its maximum and oscillated around lower values.
6. Conclusion
Against the backdrop of the rapid advancement of convolutional neural network (CNN) technologies, this study focused on the intelligent recognition of pneumonia from chest X-ray images. The research employed the AlexNet model for pneumonia classification, with the architecture extended by incorporating a flatten layer and a dropout layer. Model performance was evaluated under different epoch settings. The experimental results demonstrated that the proposed CNN-based model exhibited strong performance, achieving relatively high accuracy in distinguishing pneumonia cases from normal chest X-ray images.
Despite these promising results, several limitations remain. Specifically, the model’s generalization ability across diverse medical imaging modalities and disease categories has not yet been fully validated, leaving its broader applicability uncertain. Moreover, the study did not address small-sample scenarios, which are common in clinical practice. To adapt the model for such situations, techniques such as few-shot learning or advanced data augmentation strategies will be necessary. In addition, the overall accuracy and robustness of the model require further improvement to ensure reliable deployment in complex real-world clinical environments.
Future research should therefore focus on addressing these limitations to enhance the practical utility of the model. First, efforts should be directed toward expanding the validation scope, including testing the model across multiple imaging modalities (e.g., CT, MRI) and extending its application to other disease classification tasks, thereby enhancing adaptability in diverse clinical scenarios. Second, targeted optimization for small-sample cases can be achieved by integrating few-shot learning frameworks and advanced data augmentation techniques to sustain model performance under data-limited conditions. Finally, further optimization of the model architecture, integration of multi-source clinical data, and comprehensive robustness testing are expected to improve both accuracy and stability, ultimately laying a stronger foundation for its clinical adoption as a reliable diagnostic aid.
References
[1]. Hao, K., Hu, L., & Ding, X. (2022). Weighted Fisher convolutional neural network model for breast cancer detection. Computer Technology and Development, 32(6), 179–185.
[2]. Shen, X., Zhang, X., Wang, Q., et al. (2023). Salivary gland tumor ultrasound image classification based on convolutional neural networks. Journal of Clinical Ultrasound in Medicine, 25(10), 849–855. https: //doi.org/10.16245/j.cnki.issn1008-6978.2023.10.020
[3]. Song, X., Wang, F., & Gao, J. (2024). Sickle cell image classification model based on CNN and KNN. Journal of Mudanjiang Normal University (Natural Science Edition), (4), 22–26. https: //doi.org/10.13815/j.cnki.jmtc(ns).2024.04.013
[4]. Shao, R., Liu, J., Ma, J., et al. (2024). Review of the application of convolutional neural networks in liver cancer pathological image diagnosis. Computer Systems & Applications, 33(4), 26–38. https: //doi.org/10.15888/j.cnki.csa.009466
[5]. Liu, P., An, D., & Feng, Y. (2024). Brain tumor image segmentation based on asymmetric U-shaped convolutional neural networks. Computer Systems & Applications, 33(8), 196–204. https: //doi.org/10.15888/j.cnki.csa.009613
[6]. Wu, X., Yang, X., Li, Z., et al. (2024). Brain tumor segmentation based on a combination of U-Net and watershed algorithms. Computer and Digital Engineering, 52(9), 2764–2770.
[7]. Cao, Q., Wang, F., & Niu, J. (2020). Optimization of brain tumor medical image segmentation based on 3D convolutional neural networks. Modern Electronic Technology, 43(3), 74–77. https: //doi.org/10.16652/j.issn.1004-373x.2020.03.018
[8]. Xu, L., Li, H., Liu, Z., et al. (2024). MRI image segmentation algorithm for brain gliomas based on 3D-Ghost convolutional neural networks: 3D-GA-Unet. Journal of Computer Applications, 44(4), 1294–1302.
[9]. Chen, F., Li, M., Jiang, Y., et al. (2024). Automatic recognition and segmentation of pancreas and tumors using deep convolutional network models: Based on 3D V-Net. Journal of Molecular Imaging, 47(11), 1170–1175.
[10]. Pei, G., Zhang, S., Zhang, J., et al. (2024). ECG-UNet: A lightweight medical image segmentation algorithm based on a U-shaped structure. Journal of Applied Science, 42(6), 922–933.
[11]. Zhang, T., Zhou, D., Li, J., et al. (2024). Convolutional network for fuzzy medical image edge detection. Computer Systems & Applications, 33(2), 198–206. https: //doi.org/10.15888/j.cnki.csa.009384
[12]. Xie, D., Li, L., & Miao, C. (2021). Handwritten digit recognition based on improved AlexNet convolutional neural network. Journal of Hebei University of Engineering (Natural Science Edition), 38(4), 102–106.
[13]. Tian, Y. (2025). Development and application of convolutional neural network models. Digital Communication World, (4), 108–110.
[14]. Peng, W., Ye, Y., & Qiu, X. (2025). Clinical characteristics of pneumonia patients with different severity levels. Great Doctor, 10(5), 99–102.
Cite this article
Gao,H. (2025). Pneumonia Detection and Analysis Using AlexNet. Applied and Computational Engineering,190,1-7.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of CONF-SPML 2026 Symposium: The 2nd Neural Computing and Applications Workshop 2025
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Hao, K., Hu, L., & Ding, X. (2022). Weighted Fisher convolutional neural network model for breast cancer detection. Computer Technology and Development, 32(6), 179–185.
[2]. Shen, X., Zhang, X., Wang, Q., et al. (2023). Salivary gland tumor ultrasound image classification based on convolutional neural networks. Journal of Clinical Ultrasound in Medicine, 25(10), 849–855. https: //doi.org/10.16245/j.cnki.issn1008-6978.2023.10.020
[3]. Song, X., Wang, F., & Gao, J. (2024). Sickle cell image classification model based on CNN and KNN. Journal of Mudanjiang Normal University (Natural Science Edition), (4), 22–26. https: //doi.org/10.13815/j.cnki.jmtc(ns).2024.04.013
[4]. Shao, R., Liu, J., Ma, J., et al. (2024). Review of the application of convolutional neural networks in liver cancer pathological image diagnosis. Computer Systems & Applications, 33(4), 26–38. https: //doi.org/10.15888/j.cnki.csa.009466
[5]. Liu, P., An, D., & Feng, Y. (2024). Brain tumor image segmentation based on asymmetric U-shaped convolutional neural networks. Computer Systems & Applications, 33(8), 196–204. https: //doi.org/10.15888/j.cnki.csa.009613
[6]. Wu, X., Yang, X., Li, Z., et al. (2024). Brain tumor segmentation based on a combination of U-Net and watershed algorithms. Computer and Digital Engineering, 52(9), 2764–2770.
[7]. Cao, Q., Wang, F., & Niu, J. (2020). Optimization of brain tumor medical image segmentation based on 3D convolutional neural networks. Modern Electronic Technology, 43(3), 74–77. https: //doi.org/10.16652/j.issn.1004-373x.2020.03.018
[8]. Xu, L., Li, H., Liu, Z., et al. (2024). MRI image segmentation algorithm for brain gliomas based on 3D-Ghost convolutional neural networks: 3D-GA-Unet. Journal of Computer Applications, 44(4), 1294–1302.
[9]. Chen, F., Li, M., Jiang, Y., et al. (2024). Automatic recognition and segmentation of pancreas and tumors using deep convolutional network models: Based on 3D V-Net. Journal of Molecular Imaging, 47(11), 1170–1175.
[10]. Pei, G., Zhang, S., Zhang, J., et al. (2024). ECG-UNet: A lightweight medical image segmentation algorithm based on a U-shaped structure. Journal of Applied Science, 42(6), 922–933.
[11]. Zhang, T., Zhou, D., Li, J., et al. (2024). Convolutional network for fuzzy medical image edge detection. Computer Systems & Applications, 33(2), 198–206. https: //doi.org/10.15888/j.cnki.csa.009384
[12]. Xie, D., Li, L., & Miao, C. (2021). Handwritten digit recognition based on improved AlexNet convolutional neural network. Journal of Hebei University of Engineering (Natural Science Edition), 38(4), 102–106.
[13]. Tian, Y. (2025). Development and application of convolutional neural network models. Digital Communication World, (4), 108–110.
[14]. Peng, W., Ye, Y., & Qiu, X. (2025). Clinical characteristics of pneumonia patients with different severity levels. Great Doctor, 10(5), 99–102.









