1. Introduction
The contemporary prevalence of brain tumor underscores their significance, with fatality observed among afflicted individuals. A tumor signifies an anomalous augmentation of bodily tissues, dichotomously categorized into malignant and benign classifications [1]. Since most cancerous are hard to treat, finding tumors is really of great significance. Normally, the doctor will use imaging like computerized tomography, magnetic resonance imaging, and other ways to determine the position and the spread of the tumor [1-4]. In this case, an automated algorithm to identify the tumor from the images is possible and necessary.
There is already some research focused on automatically identifying the tumor from the images, and there are also already some successful products or models [5, 6]. For example, Govindaraj et al. provide a way to identify the tumor in the brain [5]. Nina Linder et al., also gives a way to detect the tumor automatically [6]. However, dissimilar to other artificial intelligence applications, which have numerous samples and include most cases to train the model, the size and shape of tumors for various people may differ. In this case, it is challenging to provide the model in all the possible cases to train, which may decrease the accuracy of the model, and if the model misses the tumor and the tumor is cancerous, it may affect the health.
In this regard, it is pressing for people to find a way to produce images with various views based on a few pictures to enhance the performance of image identification. One option to produce the picture is using Generative Adversarial Network (GAN). Using GAN, the model can imitate tumors of different sizes and shapes based on some tumor pictures. And this study will use these images to train a convolutional neural network (CNN) model. Then this study will compare the accuracy of CNN with the image that GAN produce and CNN without using GAN to see whether the images improve CNN’s accuracy. The experiment is focused on meningioma. During training, the model will be fed about 1595 photos with no tumor and about 1, 339 photos with meningioma. Then, the GAN model will generate 1339 images for no tumor and 1595 for meningioma so the number of total sample in two set are about the same. These images will also be used as training data for the CNN model. The testing data set includes about 405 photos that don’t have a tumor and about 306 photos with meningioma.
The rest of the paper is organized as follows: Section 2 will talk about the method the study used to build the CNN network and DCGAN network, Section 3 will discuss the result, and Section 4 will give a conclusion about this study and give some ideas for future works.
2. Method
2.1. Datasets description and preprocessing
In this study, the data is sourced from Kaggle [7]. In this dataset, there are four subsets for training and testing, which are glioma, meningioma, no tumor, and pituitary. This study will focus on identifying meningioma, so the images in the ‘meningioma’ fold and ‘no tumor’ fold are used. In these two folds, there are a total of 2934 gray-scale images with different sizes. Some sample images can be found in Figure 1.
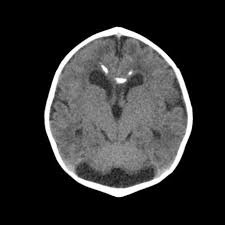
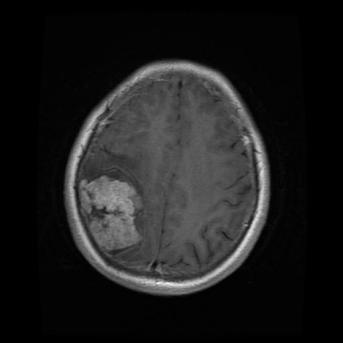
Figure 1. The sample images of the collected dataset (Left one is no tumor) [7].
In terms of the data preprocessing, this study converted all the images to RGB format and resize all the images to 256 × 256 for the DCGAN model and 512 × 512 for the CNN model. The study also does the normalization to the images.
2.2. Proposed approach
2.2.1. GAN. The goal of GANs is to estimate the potential distribution of real data samples and generate new samples from that distribution [8]. The GAN model comprises two main components: the generator and the discriminator [9]. The generator’s role is to produce a sample and make it appear as realistic as it can. On the other hand, the discriminator’s task is to determine whether an input is authentic or created by the generator [9]. The training process for the generator and discriminator occurs alternately [8].
2.2.2. DCGAN. It provides a set of convolutional neural networks, removing the pooling layer and adding Batch Normalization between the convolutional layer and the activation function to achieve local normalization, which greatly improves the network model [9]. Since DCGAN is developed based on a GAN network, it also includes a generator and discriminator with a similar purpose. DCGAN expands on GAN not only retains the ability to generate excellent data but also incorporates the advantages of CNN feature extraction, making it have an improvement in image analysis and processing capabilities [9].
2.2.3. DCGAN network construction. In this project, the model that is used to generate images is DCGAN. DCGAN is implemented by convolution neural networks [9]. In the model, there are four convolution layers in the discriminator. And there will be a batch normalization layer to normalize for the last 3 convolution layers and a LeakyRelu layer as the activation function for the all four convolution layers. There will also be a view function to reshape and a linear layer. Then return the result with a squeeze function. The generator uses Relu as the active function and BatchNorm2d to normalization for the first three transposed convolutional layers and tanh for the last transposed convolutional layer. It has linear function and view function at first. There also will be a Relu for the BatchNorm2d before all of the transposed convolution layers.
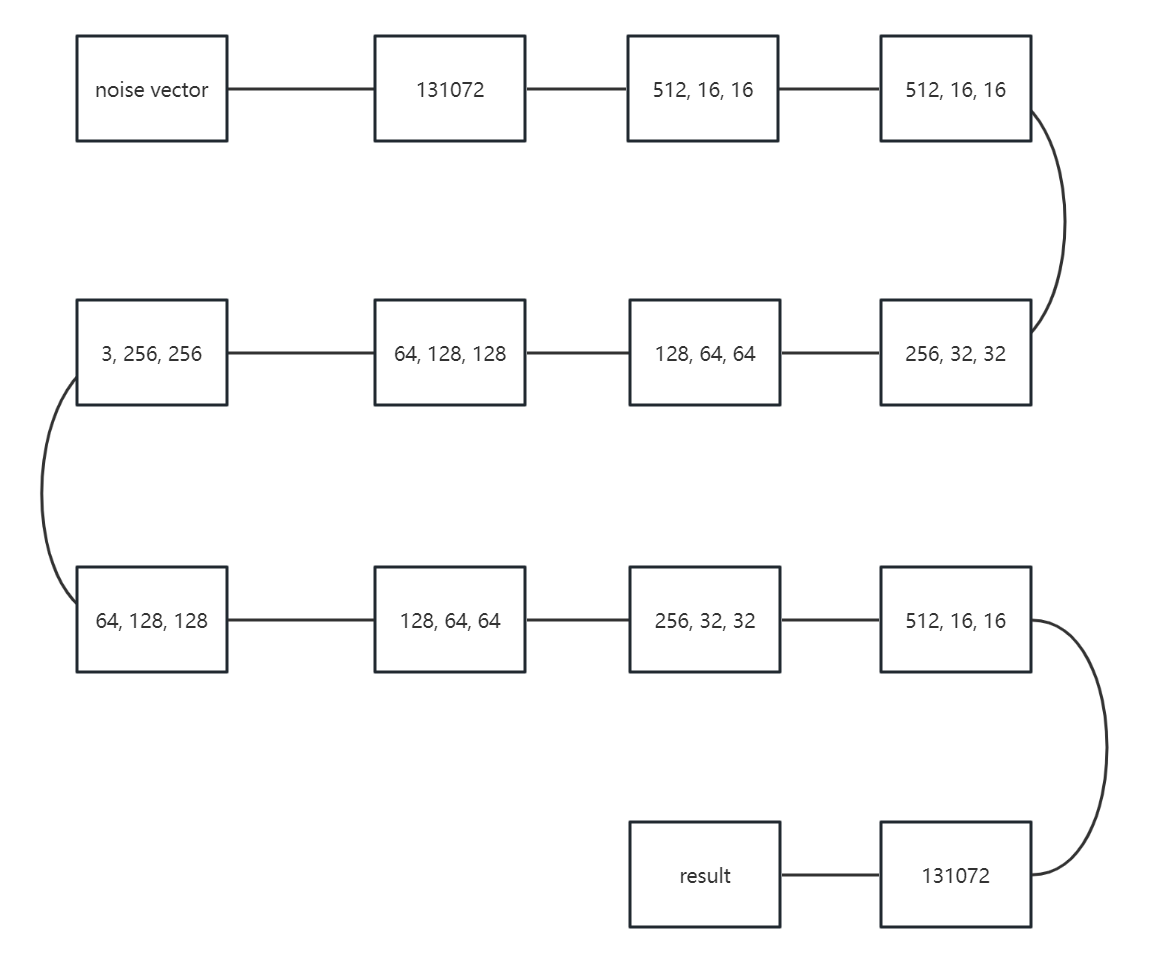
Figure 2. The structure of DCGAN.
Figure 2 is the structure of the DCGAN network. In this figure, it can be observed that the DCGAN network uses a total of thirteen steps, from receiving a noise vector to making the decision of whether it is a real image or a fake one, which is generated by the generator. The first seven steps are for the generator to generate the image, and the last six steps are for the discriminator to decide whether the image is generated by the generator or not.
2.3. Implementation details
In this project, the model needs several parameters to achieve the goal. In terms of the DCGAN model, the model has 520 epochs, which means it has been trained 520 times. The learning rate is from 0.002 to 0.00002 with cosine learning rate decay. The loss function is BCEWithLogitsLoss [10], and the batch size is 64. This project uses Adam to be the optimizer for both the generator and discriminator.
In the CNN model, three epochs is trained, with a learning rate set at 0.001 and Adam as the chosen optimizer. A batch size of 4 is utilized, and the CNN model employs distinct loss functions similar to those in the DCGAN model. The CNN model uses cross-entropy loss as the loss function. The CNN model consists of two convolution layers with the pooling layer, which uses Max Pool. Then, there will be a layer to flatten the data. The CNN model also includes three linear layers. The CNN model uses the LeakyRelu as the activation function, which is similar to the DCGAN. The study set labeled meningioma as one and no tumor as zero.
3. Results and discussion
After implementing the total of 2934 images (1595 images with meningioma and 1339 with no tumor), which are generated by DCGAN, the accuracy for the CNN model to analyze whether the image has meningioma or not is 97.75 percent. The f1 score for the CNN model is about 0.9738. This shows that the implemented images from DCGAN have improved CNN’s performance since the accuracy for the CNN model without implemented images is 93.53 percent, and the f1 score is about 0.9187. This indicates a 4.22 percent improvement in accuracy and about a 0.0551 improvement for the f1 score. Table 1 below shows the contrast of this difference.
Table 1. The Performance of Two Model.
Model | Performance | |
Accuracy | f1 score | |
CNN without DCGAN | 93.53% | 0.9187 |
CNN with DCGAN | 97.75% | 0.9738 |
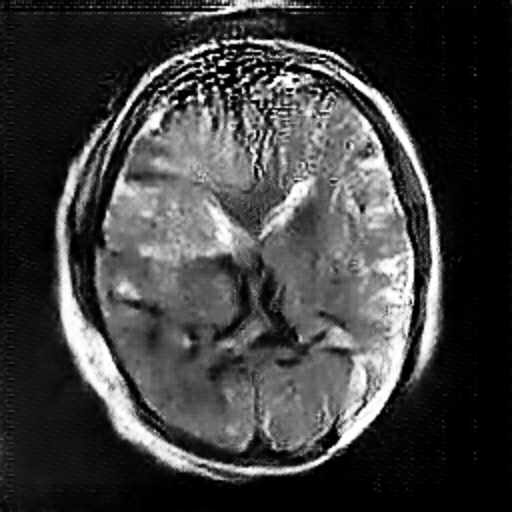
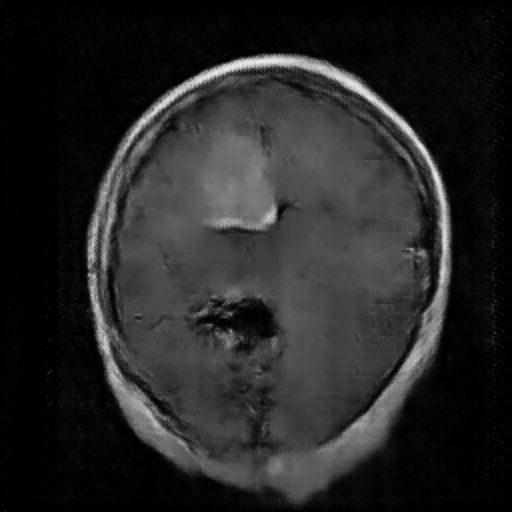
Figure 3. The Image Generated by DCGAN (Left one is no tumor).
In the image above, the right one is the image with meningioma generated by the DCGAN. We can see that there is a tumor at the upper left, which is kind of brighter. However, the complexity of the brain makes it hard for the model to learn and generate images very well. In this case, it’s not easy for the DCGAN to generate really similar images. Just like Figure 3, It rarely has the the detail for the brain compared with the original image, especially the image with meningioma. In the image with meningioma, it’s hard to find any texture or organ that should be contained in the brain. However, the CNN model may still learn from the images generated by the DCGAN by capturing specific features like lighter areas, and the CNN model has increased its accuracy.
4. Conclusion
This study is proposed to find a way to overcome the negative influence of the CNN network to identify the images that have meningioma or not, which is given by the shortage of training data. The study used the DCGAN network to generate images that have meningioma or no tumor. Utilizing both the generated images and the original ones in tandem, the CNN network is trained with the objective of enhancing its ability to classify two distinct image types. The experimental outcomes demonstrate that, in contrast to exclusively using the original dataset for CNN training, the inclusion of DCGAN-generated images leads to an enhancement in the accuracy of the CNN network's predictions. Looking ahead, there is a prospect of generating images at a larger size of 512 × 512 and concurrently refining the neural network architecture. Alternatively, images sized at 256 × 256 could be employed for both DCGAN and CNN. This consideration arises from the fact that resizing images can result in the loss or alteration of certain details. We can also apply DCGAN for the specific aspect image like overlook or side-looking and mix the generated image. Maybe this way can eliminate the disturbance by different aspect images.
References
[1]. MedlinePlus 2022 https://medlineplus.gov/ency/article/001310.htm
[2]. Deepak S and Ameer P M 2019 Brain tumor classification using deep CNN features via transfer learning. Computers in biology and medicine 111 103345
[3]. Qiu Y Chang C S Yan J L Ko L and Chang T S 2019 Semantic segmentation of intracranial hemorrhages in head CT scans In 2019 IEEE 10th International Conference on Software Engineering and Service Science (ICSESS) pp 112-115 IEEE
[4]. Kaldera H N T K et al 2019 March Brain tumor classification and segmentation using faster R-CNN In 2019 Advances in Science and Engineering Technology International Conferences (ASET) pp 1-6 IEEE
[5]. Govindaraj V and Murugan P R 2014 A complete automated algorithm for segmentation of tissues and identification of tumor region in T1, T2, and FLAIR brain images using optimization and clustering techniques International journal of imaging systems and technology 24(4) pp 313-325
[6]. Linder N et al 2012 Identification of tumor epithelium and stroma in tissue microarrays using texture analysis Diagnostic pathology 7 pp 1-11
[7]. Msoud N 2021 Brain Tumor MRI Dataset Kaggle https://www.kaggle.com/datasets/masoudnickparvar/brain-tumor-mri-dataset
[8]. Wang K Gou C Duan Y Lin Y Zheng X and Wang F Y 2017 Generative adversarial networks: introduction and outlook IEEE/CAA Journal of Automatica Sinica 4(4) pp 588-598
[9]. Fang W Zhang F Sheng V S and Ding Y 2018 A Method for Improving CNN-Based Image Recognition Using DCGAN Computers Materials & Continua 57(1)
[10]. Sapora S Lazarescu B & Lolov C 2019 Absit invidia verbo: Comparing deep learning methods for offensive language. arXiv preprint arXiv:1903.05929.
Cite this article
Chen,M. (2024). DCGAN-based data augmentation for improving CNN performance in meningioma classification. Applied and Computational Engineering,51,1-5.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 4th International Conference on Signal Processing and Machine Learning
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. MedlinePlus 2022 https://medlineplus.gov/ency/article/001310.htm
[2]. Deepak S and Ameer P M 2019 Brain tumor classification using deep CNN features via transfer learning. Computers in biology and medicine 111 103345
[3]. Qiu Y Chang C S Yan J L Ko L and Chang T S 2019 Semantic segmentation of intracranial hemorrhages in head CT scans In 2019 IEEE 10th International Conference on Software Engineering and Service Science (ICSESS) pp 112-115 IEEE
[4]. Kaldera H N T K et al 2019 March Brain tumor classification and segmentation using faster R-CNN In 2019 Advances in Science and Engineering Technology International Conferences (ASET) pp 1-6 IEEE
[5]. Govindaraj V and Murugan P R 2014 A complete automated algorithm for segmentation of tissues and identification of tumor region in T1, T2, and FLAIR brain images using optimization and clustering techniques International journal of imaging systems and technology 24(4) pp 313-325
[6]. Linder N et al 2012 Identification of tumor epithelium and stroma in tissue microarrays using texture analysis Diagnostic pathology 7 pp 1-11
[7]. Msoud N 2021 Brain Tumor MRI Dataset Kaggle https://www.kaggle.com/datasets/masoudnickparvar/brain-tumor-mri-dataset
[8]. Wang K Gou C Duan Y Lin Y Zheng X and Wang F Y 2017 Generative adversarial networks: introduction and outlook IEEE/CAA Journal of Automatica Sinica 4(4) pp 588-598
[9]. Fang W Zhang F Sheng V S and Ding Y 2018 A Method for Improving CNN-Based Image Recognition Using DCGAN Computers Materials & Continua 57(1)
[10]. Sapora S Lazarescu B & Lolov C 2019 Absit invidia verbo: Comparing deep learning methods for offensive language. arXiv preprint arXiv:1903.05929.









