1. Introduction
Lung X-ray image segmentation is one of the important research directions in the field of medical image processing, and its main purpose is to differentiate and segment lung tissues from other tissues (e.g., heart, bones, etc.) in X-ray images, so as to help doctors accurately diagnose diseases. Lung X-ray images face many challenges in the segmentation process of traditional image processing methods due to their complex structure and grey scale changes, so the application of deep learning algorithms has brought revolutionary breakthroughs in lung X-ray image segmentation in recent years.
Deep learning algorithms, as a cutting-edge technology in the field of artificial intelligence, are of great significance in lung X-ray image segmentation [1]. Firstly, deep learning algorithms are able to learn complex feature representations through a large amount of data, which improves the recognition of lung structures. Secondly, deep learning algorithms have strong nonlinear fitting ability, which can better adapt to the irregular shapes and large grey scale changes in lung X-ray images [2,3]. In addition, the deep learning algorithm is able to automatically learn the feature representation, reducing the reliance on manually designed features and improving the accuracy and stability of the segmentation results.
In the lung X-ray image segmentation task, commonly used deep learning models include U-Net [4], SegNet [5], DeepLab [6], and so on. These models achieve accurate segmentation of lung tissues while preserving spatial information by building deep neural network structures [7]. Meanwhile, for different types of lung diseases (e.g., tumours, infections, etc.), the network structure and loss function can also be adjusted in a targeted manner to further improve the segmentation effect.
In conclusion, deep learning algorithms play an important role in lung X-ray image segmentation and bring great impetus to improve the efficiency, accuracy, and clinical diagnosis of medical image processing. In this paper, based on the traditional Unet model, we have carried out structural innovation and improvement, integrated the latest GSConv module to optimise and improve the Unet, and compared the segmentation effect of the two methods, which provides a way of thinking for lung X-ray image segmentation.
2. Data set sources
The dataset chosen in this paper is an open-source dataset, the dataset is selected from the open-source database, the database contains 150 lung X-ray images and their corresponding 150 masks, and we select four of them for presentation, the results are shown as follows, the four images in the first row are the original images of the lung X-ray, and the images in the second row in the corresponding position are their corresponding masks, as shown in Fig. 1.
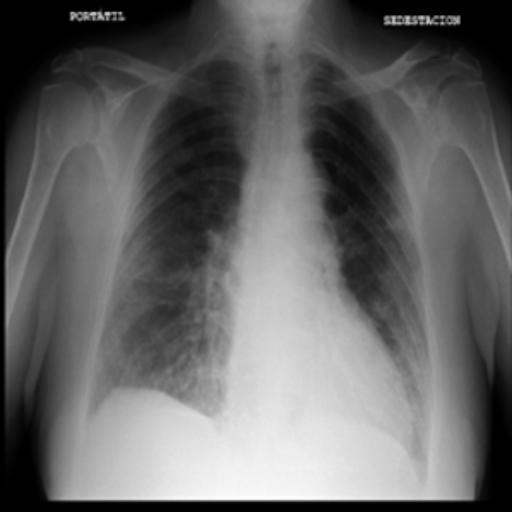
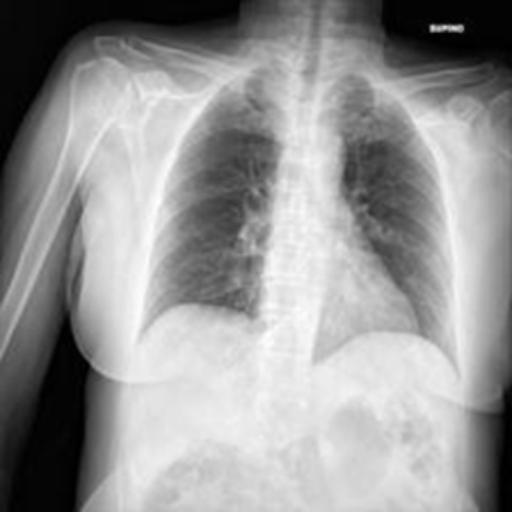
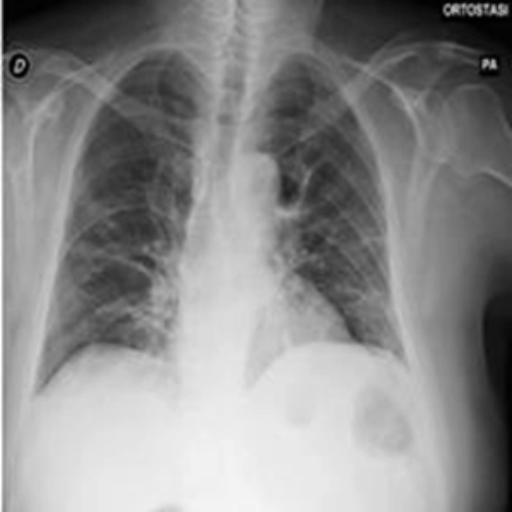
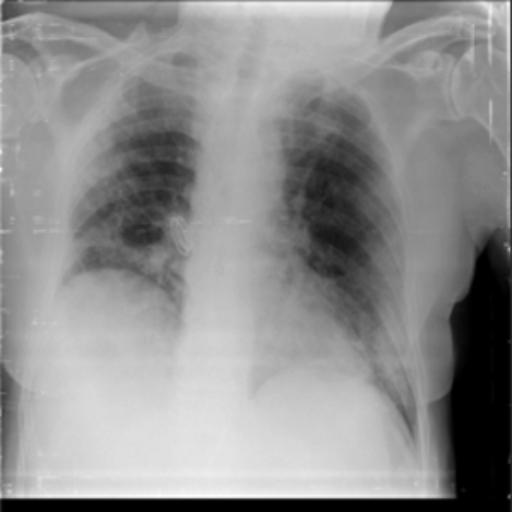




Figure 1. Partial data.
3. Method
3.1. Unet
The U-Net model is a classical deep-learning network structure originally proposed by Ronneberger et al. in 2015, specifically for image segmentation tasks. The design of U-Net is inspired by the full convolutional network (FCN) and the encoder-decoder structure, which has strong feature extraction and spatial information retention capabilities [8].
The U-Net model is mainly divided into two parts: encoder and decoder. The encoder is responsible for gradually extracting the features of the input image through convolutional and pooling layers and compressing the spatial information. Subsequently, the decoder gradually recovers the size of the feature map through upsampling operations and combines the high-level semantic information extracted in the encoder with the underlying detail information to achieve accurate segmentation results. In addition, U-Net introduces a jump connection mechanism, i.e., the feature map of a layer in the encoder is directly connected to the corresponding layer in the decoder, which helps to overcome the gradient vanishing problem and improve the segmentation effect.
3.2. Graph Structure Convolution
Graph Structure Convolution (GSConv) module is a key component in a type of convolutional neural network for image segmentation tasks, designed to process data with graph structure, the GSConv module is mainly used to process non-regular image data and is able to capture spatial information and relationships between features in an image [9].
The principle of the GSConv module is based on the Graph Convolutional Network (GCN), which enables effective feature extraction and information transfer by modelling the graph structure. In GSConv, each node represents a pixel point or region in an image, and the connections between nodes indicate the spatial relationships between them. By learning the adjacency matrix and feature representations between nodes, GSConv can perform effective feature aggregation and updating while preserving local and global information. The schematic diagram of the GSConv module is shown in Fig. 2.
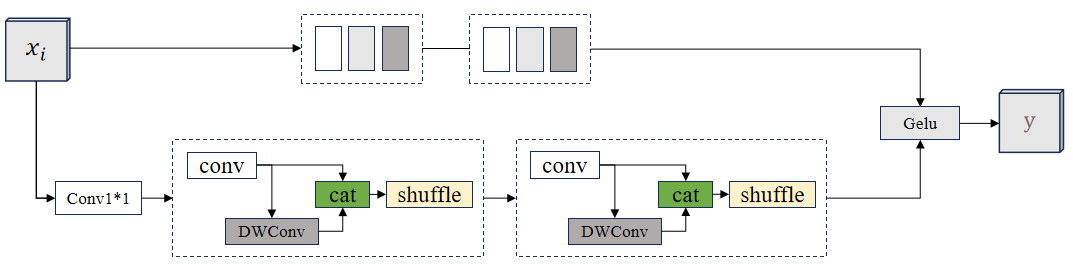
Figure 2. The schematic diagram of the GSConv module.
(Photo credit : Original)
The GSConv module processes images by first calculating the neighbourhood matrix between nodes to describe the relationship between them, then applying a convolution operation at each node to extract features, followed by aggregating and updating the features using the neighbourhood matrix, and finally, combining operations such as pooling or upsampling to achieve multi-scale feature fusion and information transfer [10].
3.3. GSConv U-Net
In order to improve the U-Net model and incorporate the GSConv module to better handle the image segmentation task, we integrated the GSConv module into the encoder and decoder parts of U-Net to take advantage of its feature extraction and information transfer capabilities for image data. The structure of the improved GSConv U-Net model is shown in Figure 3.
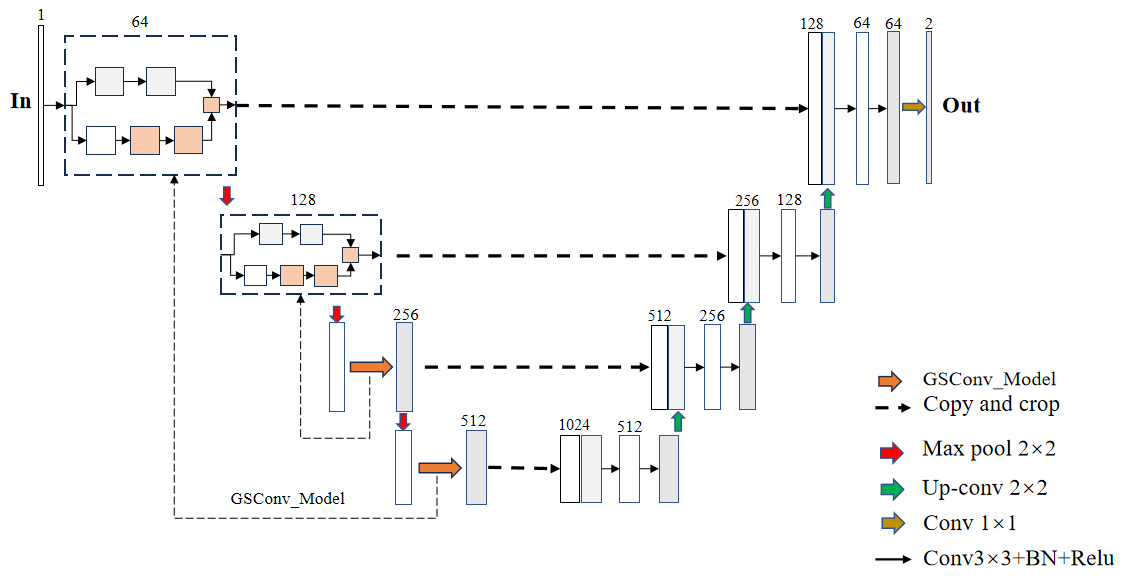
Figure 3. The structure of the improved GSConv U-Net model.
(Photo credit : Original)
Integrating GSConv into U-Net encoders: In the U-Net encoder, we replace the traditional convolutional layer with the GSConv module. This allows the GSConv module to better capture the spatial information and relationships between features in the image, thus improving the quality of feature representation and the fusion of multi-scale information.
Integration of GSConv into the U-Net decoder section: In the decoder of U-Net, the GSConv module is also added for aggregating and updating features during the up-sampling process. By introducing the GSConv module in the decoder section, it can help the network to better recover detailed information and achieve accurate segmentation results.
4. Result
Pre-processing of the image before the start of the experiment, firstly, the image size is adjusted, uniformly adjusted to the size of 512 × 512 sizes, secondly, the image is processed, and all the images are converted to a greyscale map, and finally, the image is enhanced, here we used histogram equalisation to enhance the image to facilitate the later classification.
In the experimental setup, we used the experimental equipment with 32G memory and 4090 graphics card for the experiment, epoch was set to 50, Unet and GSConv Unet models were referenced for training, the learning rate was set to 0.0002, the batch size was set to 32, and the ratio of the training set, validation set, and test set was set to 4:4:3, the experiment was conducted in the pytorch framework and python version 3.9, the changes of the loss values of the training set and validation set are recorded during the experiment, and the evaluation parameters such as ACCURACY and IOU of the test set are outputted at the end of the experiment.
Firstly, we output the change of the loss of the training set and validation set during the training process, as shown in Fig. 4 for the Unet model training set and validation set, and as shown in Fig. 5 for the GSConv Unet model training set and validation set, with the red colour denoting the loss of the training set and the orange colour denoting the loss of the validation set, and the curves denoting the curves after smoothing.
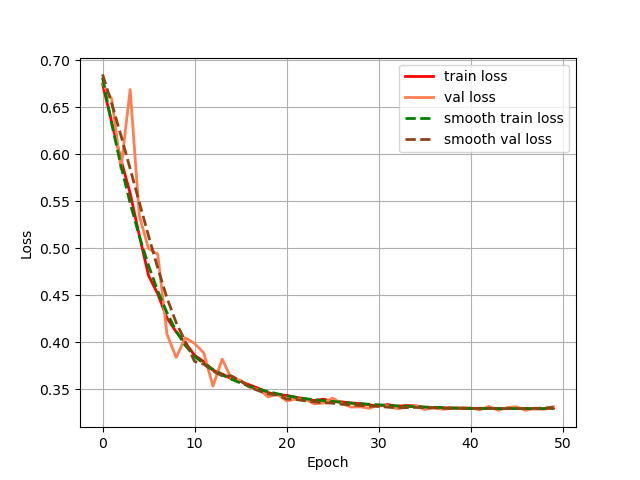
Figure 4. The change of the loss of the training set.
(Photo credit : Original)
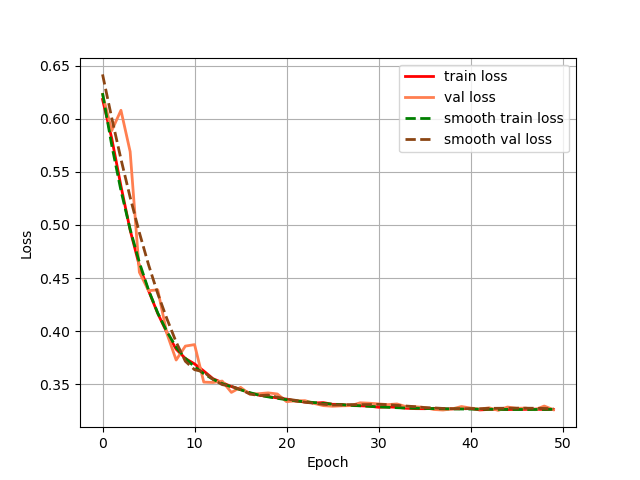
Figure 5. The change of the loss of the validation set.
(Photo credit : Original)
From the loss change curves of Unet and GSConv Unet, it can be seen that GSConv Unet converges faster and reaches smaller loss values earlier, and the loss values of the GSConv Unet training and validation sets almost overlap, which indicates that the model's generalisation ability is better, and the two models converge in the end.
In the testing part, the evaluation indexes of the two models of Unet and GSConv Unet are outputted respectively, and GSConv Unet is 9.01% higher than Unet in segmentation accuracy, and 9.76 higher than Unet in iou, and the segmentation results of Unet and GSConv Unet are outputted as shown in Fig. 6, with the first column being the lung X-ray image, the second column is the mask of the golden segmentation standard, the third column is the segmentation result of the Unet model, and the fourth column is the segmentation result of the GSConv Unet model.
Table 1. Modelling assessment.
Model | Accuracy | Miou |
Unet | 81.46% | 77.15 |
GSConv Unet | 90.47% | 86.91 |
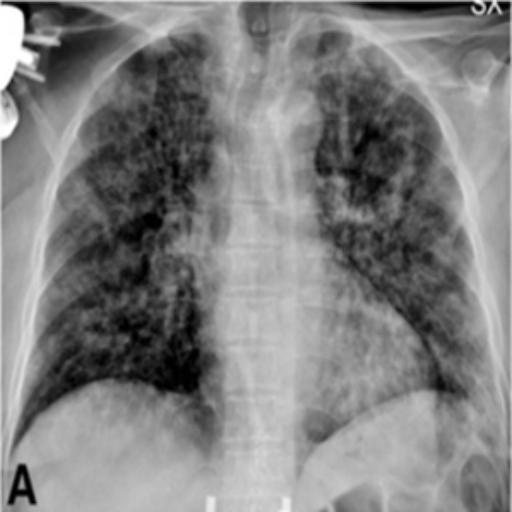
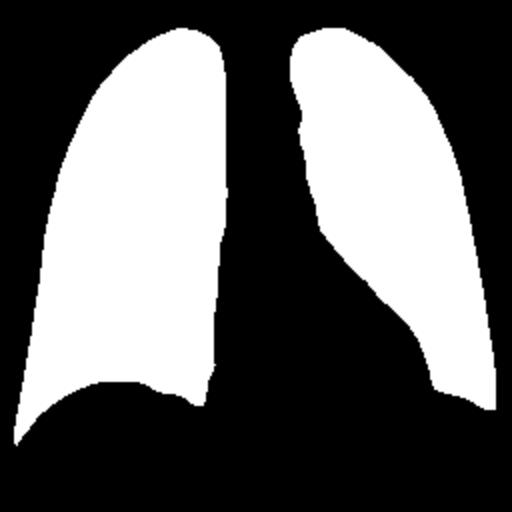

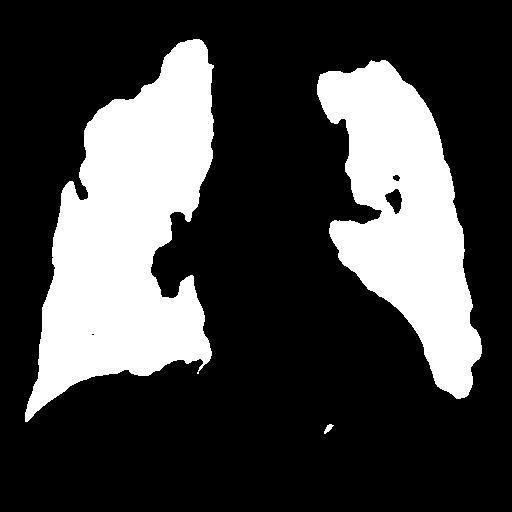
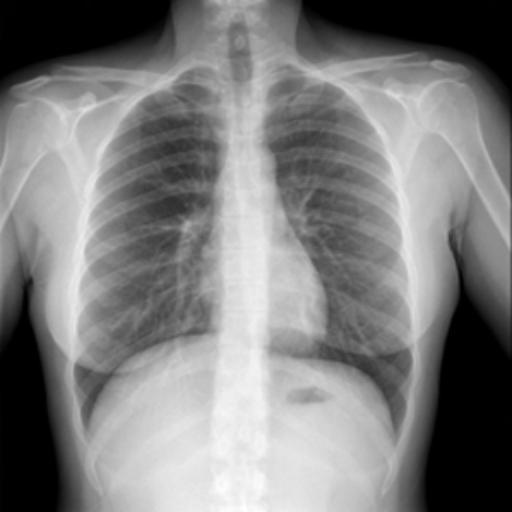



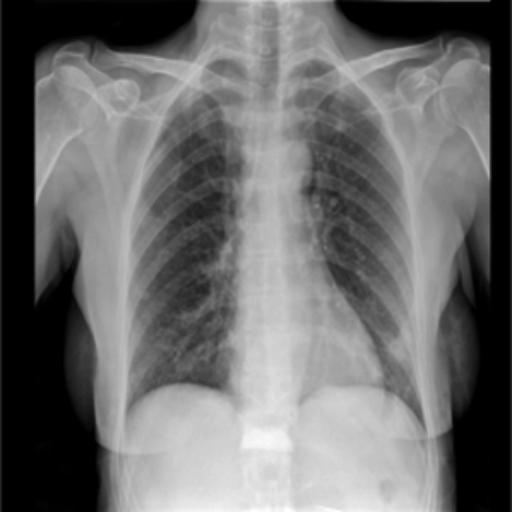

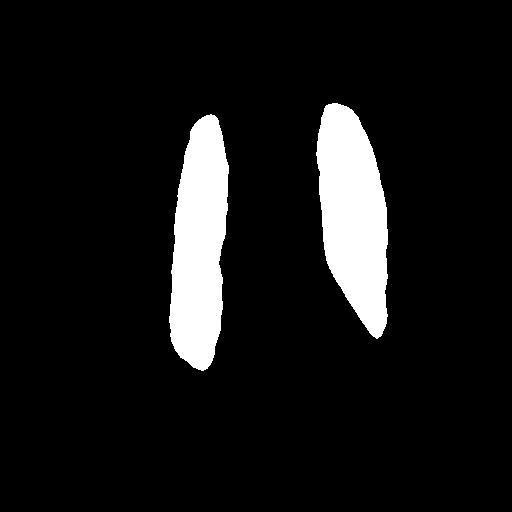

Figure 6. Test Set Output Results.
(Photo credit : Original)
From the segmentation results of the Unet and GSConv Unet models, GSConv Unet is more accurate and more detailed in segmenting the lungs compared to Unet, and on balance, GSConv Unet has a significant improvement compared to Unet.
5. Conclusion
In this paper, we optimise Unet by innovating and improving the structure of the traditional Unet model, introducing the latest GSConv module, and comparing the segmentation effect of the two methods, which provides new ideas for lung X-ray image segmentation. In the process of optimising the Unet model and integrating the GSConv module to handle the image segmentation task more efficiently, we incorporate the GSConv module into the encoder and decoder parts of U-Net in order to make full use of its feature extraction and information transfer capabilities. Before the experiment, the images were first resized to a uniform size of 512×512, and then all the images were converted to grey-scale maps and enhanced by applying histogram equalisation. The Unet and GSConv Unet models are used in the training phase, and it can be observed from the loss change curves that GSConv Unet converges faster and reaches smaller loss values early; at the same time, the losses on the training and validation sets almost overlap, which indicates that GSConv Unet has a better generalisation ability. Both models eventually converge. In the testing stage, the evaluation index outputs of both models, Unet and GSConv Unet, were performed, and GSConv Unet was 9.01% and 9.76% higher in segmentation accuracy and iou, respectively, which showed that GSConv Unet depicted the lung contour more accurately and in more detail compared to Unet, as shown by the segmentation results. Taken together, GSConv Unet is significantly improved compared to Unet.
In summary, the structurally innovative and improved GSConv Unet model in this study exhibits faster convergence speed, better generalisation ability, and higher segmentation accuracy and iou metrics compared to traditional Unet. This research result explores new paths for the medical image processing field and provides useful insights for future research in related fields.
References
[1]. Sulaiman, Adel, et al. "A convolutional neural network architecture for segmentation of lung diseases using chest X-ray images." Diagnostics 13.9 (2023): 1651.
[2]. Ullah, Ihsan, et al. "A deep learning based dual encoder-decoder framework for anatomical structure segmentation in chest X-ray images." Scientific Reports 13.1 (2023): 791.
[3]. de Almeida, Pedro Aurélio Coelho, and Díbio Leandro Borges. "A deep unsupervised saliency model for lung segmentation in chest X-ray images." Biomedical Signal Processing and Control 86 (2023): 105334.
[4]. Arvind, S., et al. "Improvised light weight deep CNN based U-Net for the semantic segmentation of lungs from chest X-rays." Results in Engineering 17 (2023): 100929.
[5]. Öztürk, Şaban, and Tolga Çukur. "Focal modulation network for lung segmentation in chest X-ray images." Turkish Journal of Electrical Engineering and Computer Sciences 31.6 (2023): 1006-1020.
[6]. Ghimire, Samip, and Santosh Subedi. "Estimating Lung Volume Capacity from X-ray Images Using Deep Learning." Quantum Beam Science 8.2 (2024): 11.
[7]. Gaggion, Nicolás, et al. "CheXmask: a large-scale dataset of anatomical segmentation masks for multi-center chest x-ray images." Scientific Data 11.1 (2024): 511.
[8]. Brioso, Ricardo Coimbra, et al. "Semi-supervised multi-structure segmentation in chest X-ray imaging." 2023 IEEE 36th International Symposium on Computer-Based Medical Systems (CBMS). IEEE, 2023.
[9]. Chen, Lingdong, et al. "Development of lung segmentation method in x-ray images of children based on TransResUNet." Frontiers in radiology 3 (2023): 1190745.
[10]. Iqbal, Ahmed, Muhammad Usman, and Zohair Ahmed. "Tuberculosis chest X-ray detection using CNN-based hybrid segmentation and classification approach." Biomedical Signal Processing and Control 84 (2023): 104667.
Cite this article
Yang,R. (2024). Lung X-ray image segmentation based on improved Unet deep learning network algorithm with GSConv module. Applied and Computational Engineering,69,43-48.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 6th International Conference on Computing and Data Science
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Sulaiman, Adel, et al. "A convolutional neural network architecture for segmentation of lung diseases using chest X-ray images." Diagnostics 13.9 (2023): 1651.
[2]. Ullah, Ihsan, et al. "A deep learning based dual encoder-decoder framework for anatomical structure segmentation in chest X-ray images." Scientific Reports 13.1 (2023): 791.
[3]. de Almeida, Pedro Aurélio Coelho, and Díbio Leandro Borges. "A deep unsupervised saliency model for lung segmentation in chest X-ray images." Biomedical Signal Processing and Control 86 (2023): 105334.
[4]. Arvind, S., et al. "Improvised light weight deep CNN based U-Net for the semantic segmentation of lungs from chest X-rays." Results in Engineering 17 (2023): 100929.
[5]. Öztürk, Şaban, and Tolga Çukur. "Focal modulation network for lung segmentation in chest X-ray images." Turkish Journal of Electrical Engineering and Computer Sciences 31.6 (2023): 1006-1020.
[6]. Ghimire, Samip, and Santosh Subedi. "Estimating Lung Volume Capacity from X-ray Images Using Deep Learning." Quantum Beam Science 8.2 (2024): 11.
[7]. Gaggion, Nicolás, et al. "CheXmask: a large-scale dataset of anatomical segmentation masks for multi-center chest x-ray images." Scientific Data 11.1 (2024): 511.
[8]. Brioso, Ricardo Coimbra, et al. "Semi-supervised multi-structure segmentation in chest X-ray imaging." 2023 IEEE 36th International Symposium on Computer-Based Medical Systems (CBMS). IEEE, 2023.
[9]. Chen, Lingdong, et al. "Development of lung segmentation method in x-ray images of children based on TransResUNet." Frontiers in radiology 3 (2023): 1190745.
[10]. Iqbal, Ahmed, Muhammad Usman, and Zohair Ahmed. "Tuberculosis chest X-ray detection using CNN-based hybrid segmentation and classification approach." Biomedical Signal Processing and Control 84 (2023): 104667.









