1. Introduction
Inattention, hyperactivity, and impulsive behavior are hallmarks of attention deficit hyperactivity disorder (ADHD), a neurodevelopmental disorder that causes significant disruption to daily functioning [1]. The likelihood of ADHD persisting into adulthood generally falls within the range of 4% to 5% [2], with affected individuals more likely to experience the onset of mental health conditions such major depressive disorder, bipolar disorder, and drug addiction [3]. Therefore, the precise and timely diagnosis of ADHD holds paramount importance for effective intervention and management. However, traditional ADHD diagnosis methods face challenges stemming from variability in subjective assessments, inaccuracies or incompleteness in evaluation questionnaires, and cultural factors not adequately considered in standardized ADHD tests [4]. Given these limitations, novel approaches utilizing neuroimaging techniques and artificial intelligence (AI) have emerged as promising avenues for improving ADHD diagnosis [5].
In 2011, the ADHD-200 Consortium held an international competition and made available a dataset of eight different independent neuroimaging scanning sites' resting state fMRI [6]. Functional magnetic resonance imaging (fMRI) can be utilized in ADHD categorization by recording the functional activity of the brain, which may vary between persons with ADHD and those without [7]. Thus, using machine learning (ML) and deep learning (DL) algorithms to neuroimaging data allows for the identification of complex patterns and characteristics associated with ADHD disease.
Early approaches to ADHD classification predominantly relied on machine learning techniques. Originally, these tactics centered on selecting neurobiologically significant components from input data and inputting them into sequential classifiers [8]. Their ADHD classification approaches include Attributed Graph Distance Measure [9], Support Vector Machine with Recursive Feature Elimination (SVM-RFE) [10], and Clustering [11]. To enhance classification accuracy, numerous machine learning studies have concentrated on acquiring more reliable information about ADHD by extracting latent features. For example, Fusion fMRI [12] combines Elastic Net-based feature choice with clustering methods to extract discriminative features from both non-imaging and dense functional brain networks. Subsequently, a Support Vector Machine classifier is trained to classify ADHD against control subjects. L1BioSVM [13] presents a bi-objective approach to ADHD classification, utilizing the L1-norm support vector machine (SVM) to consider both the margin of separation and empirical error simultaneously. This method applies the normal boundary intersection (NBI) technique to generate a representative nondominated set. The R-Relief method [14] employs Principal Component Analysis (PCA) to calculate sequential entropy for the score-form fractional Amplitude of Low-Frequency Fluctuation (fALFF), successfully reducing noise interference within the input data and generating resilient components for the classifier.
This shift towards DL approaches in recent years marks a departure from earlier ML methods, employing a range of neural network architectures to acquire customizable high-level features and steer the classification process. Convolutional neural networks (CNNs) have emerged as a prominent option in this domain. For examining the local spatial patterns of MRI characteristics, both 3-D CNN [15] and 4-D CNN [16] architectures have been introduced. DeepFMRI[17] is an end-to-end learning system designed for fMRI data processing. It consists of three networks: a feature extractor, a functional connection network, and a classification network. The goal is to automate the designation of participants as ADHD or healthy controls. Transformer models, known for their remarkable performance in various tasks, have also been applied to ADHD classification. Transformer with a Diffusion Kernel Attention Network is proposed that utilizes for integrated modeling of functional brain networks [18]. Autoencoder (AE) networks excel at learning discriminative, high-level features. The STAAE framework [19] decomposes rfMRI into spatial and temporal patterns using autoencoder (AE) networks, and introduces a resting state temporal template (RSTT)-based classification technique that has been validated for ADHD.
These innovative approaches signify progress in utilizing advanced techniques in neuroimaging and AI to enhance the precision of ADHD diagnosis. By leveraging these technologies, there is potential to improve intervention and management strategies for individuals with ADHD, ultimately leading to better outcomes. However, these methodologies face challenges. They lack consideration regarding whether the extracted features truly represent the primary characteristics of the input images.
To address these challenges, this study proposes a novel framework integrating the Convolutional Block Attention Module (CBAM) [20] with an autoencoder network for precise ADHD classification using fMRI data. The proposed method enhances diagnostic accuracy by leveraging CBAM to focus on salient features within fMRI scans and employing image reconstruction techniques to enhance feature representations. This framework simplifies model complexity by eliminating the decoder after model training and retaining only the encoder and classifier layers for prediction. This adjustment enhances its compatibility for deployment on mobile devices and streamlines the retraining process.
In this research, the architecture of the proposed CBAM-autoencoder network is presented, elucidating its constituent elements and operational principle, and then the design and implementation of the proposed CBAM-autoencoder network are presented, along with an evaluation of its performance on the ADHD-200 dataset. The experiments conducted reveal consistent enhancements in classification accuracy compared to existing methodologies, underscoring the effectiveness of attention-based feature extraction in ADHD diagnosis.
This paper has the following three academic contributions:
- A novel autoencoder neural network integrating attention mechanisms architecture is proposed, improving classification accuracy on the ADHD-200 dataset.
- Post-model training, the decoder is discarded, leaving only the encoder and classification layers for prediction, thereby optimizing model efficiency.
- Employing CBAM enhances the network's capability to concentrate on crucial features while disregarding irrelevant ones.
2. Proposed Methodology
2.1. Overall Architecture
The proposed method for accurate classification of Attention Deficit Hyperactivity Disorder (ADHD) using resting-state functional Magnetic Resonance Imaging (fMRI) data features a CBAM-autoencoder network (refer to Figure 1). This network harnesses the power of autoencoders to compress input data into a latent-space representation, which is then reconstructed to produce the output. The Convolutional Block Attention Module (CBAM) is a powerful mechanism used in neural networks, particularly in architectures like autoencoders, to selectively focus on important features while suppressing irrelevant ones in input images [20].
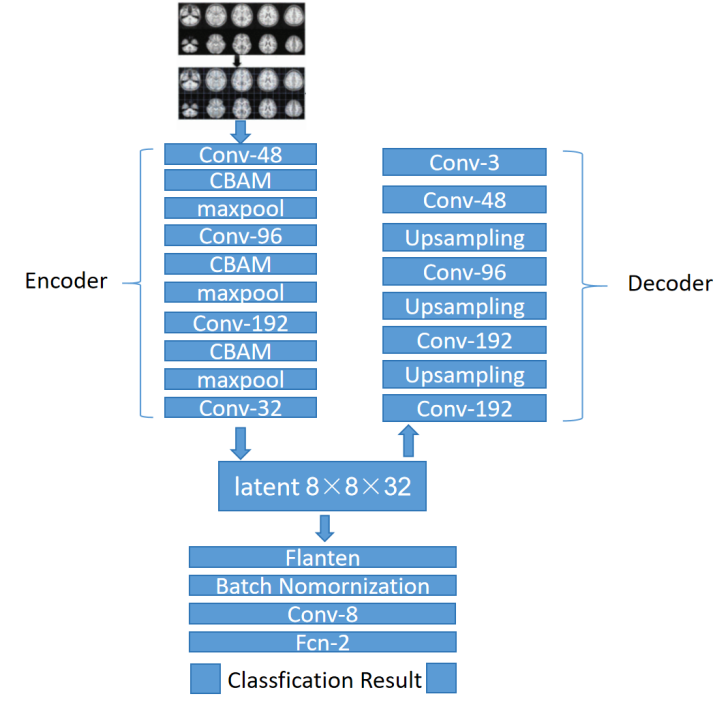 Figure 1. Overall architecture of CBAM-autoencoder network
Figure 1. Overall architecture of CBAM-autoencoder network
2.2. Autoencoder Network
The autoencoder network is made up of two components: an encoder and decoder. The encoder reduces the dimensionality of the input fMRI images (64×64×3) to generate a latent-space representation (8×8×32), while the decoder reconstructs the original input from this representation (256×256×3). It includes convolutional layers, a CBAM module, and a pooling layer. The latent-space representation is further processed by convolutional layers before reaching the classification layer. The involved layers are described in the following:
Convolutional Layer, at the heart of the autoencoder network lies the convolutional layer, which extracts features from the input images. By convolving the input data with learnable filters, this layer captures hierarchical features crucial for classification. The size of the receptive field influences the breadth of information captured, as larger fields tend to encompass more global and semantic-level details.
Batch Normalization Layers, to accelerate network training and improve stability, batch normalization layers are introduced. They normalize the input of each layer by adjusting mean and variance, enabling higher learning rates and reducing training time. Additionally, they provide regularization and improve network accuracy by alleviating the need for careful parameter initialization.
The Convolutional Block Attention Module (CBAM) is described in depth in Section 2.3.
Max-Pooling Layer, inserted between convolutional layers, the max-pooling layer reduces feature map dimensionality, compressing features and simplifying network complexity. It improves robustness and reduces overfitting by eliminating non-maximum values and achieving translation invariance.
UpSampling Layer, in the decoder, the upsampling layer restores the latent-space representation to the original image size using interpolation. Alongside convolutional layers, it converts the low-dimensional representation back to the original image format, facilitating accurate reconstruction.
2.3. Convolutional Block Attention Module
The CBAM module (see Figure 2) consists of both channel-wise and spatial-wise attention processes, each serving a specific purpose in enhancing the network's ability to capture meaningful information [20]. The channel attention module evaluates the significance of features across different channels, while the spatial attention module identifies crucial regions within feature maps. Integrating the CBAM module following the convolutional layer enables the autoencoder network to acquire and emphasize important features present in input images.
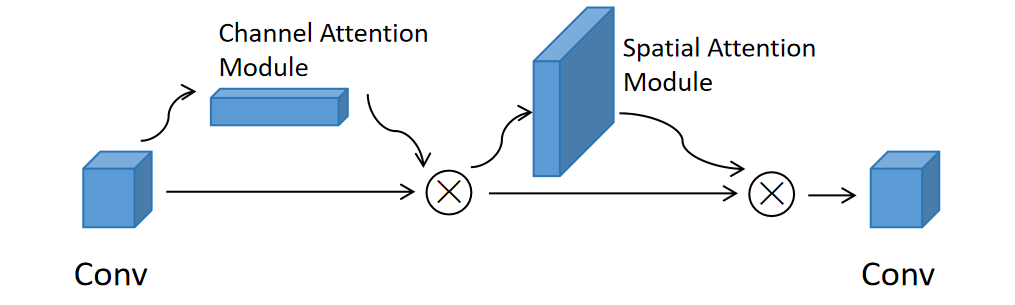
Figure 2. The CBAM module
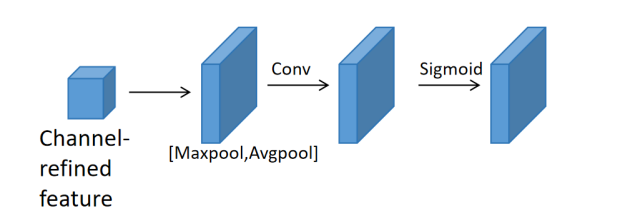
Figure 3. The channel attention module
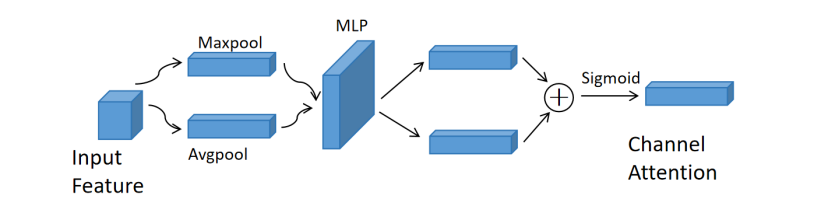
Figure 4. The spatial attention module
When an intermediate feature map \( {F∈R^{C×H×W}} \) is fed into the Convolutional Block Attention Module (CBAM), it undergoes a sequential process resulting in two crucial outputs: a 1D channel attention map \( {{M_{c}}∈R^{C×1×1}} \) and a 2D spatial attention map \( {{M_{s}}∈R^{1×H×W}} \) . This attention mechanism can be summarized as follows:
\( {F_{0}}={M_{c}}(F)⨀F \) (1)
\( {F_{00}}={M_{s}}({F_{0}})⨀{F_{0}} \) (2)
where \( ⨀ \) denotes element-wise multiplication.
The Channel Attention Module (see Figure 3) is adept at discerning the relationships between channels to generate a detailed channel attention map. This process involves compressing the spatial dimension of the input feature map, followed by aggregating spatial information through both average-pooling and max-pooling operations. By doing so, the module effectively gathers essential cues regarding distinctive object features.
\( {M_{c}}(F)=σ(MLP(AvgPool(F))+MLP(MaxPool(F))) \) (3)
where \( σ \) denotes the sigmoid function, and \( MLP \) represents a multi-layer perceptron with one hidden layer.
Contrarily, the Spatial Attention Module (see Figure 4) directs its focus towards the spatial intricacies within features. It excels in identifying informative spatial regions. The module first performs average-pooling and max-pooling procedures down a channel line to compute spatial attention, which highlights important locations. The outputs of these operations are then concatenated to form a concise feature descriptor. The spatial attention map is obtained by passing this explanation through an average layer of convolution for additional processing. This map offers guidance on where to accentuate or diminish features, thereby enhancing the feature representation.
Expanding on the process, the channel information within the feature map undergoes aggregation through both average-pooling and max-pooling operations. This aggregation produces two distinct 2D maps: \( {{{F_{s}}^{avg}}∈R^{1×H×W}} \) and \( {{{F_{s}}^{max}}∈R^{1×H×W}} \) , representing the average-pooled and max-pooled features across the channel axis, respectively. These maps capture essential insights into the feature distribution.
Following this, the two maps are concatenated to form a unified representation, incorporating both the average and maximum pooled features. Subsequently, this concatenated representation is subjected to a 7x7 convolutional operation denoted as
\( {M_{s}}(F)=σ({f_{7x7}}([AvgPool(F);MaxPool(F)]))=σ({f_{7x7}}([{{F_{s}}^{avg}};{{F_{s}}^{max}}])) \) (4)
where \( σ \) represents the sigmoid function and \( {f_{7x7}} \) denotes a convolution operation with a 7x7 filter size.
3. Experiment
3.1. Dataset
The fMRI data utilized in this investigation is sourced from the ADHD-200 repository, renowned for providing an extensive dataset catering to both competitive and research endeavors. This dataset encompasses a total of 776 fMRI scans paired with T1-weighted structural scans. Among these, 491 scans stem from typically developing individuals, while 285 scans originate from patients diagnosed with ADHD, spanning an age range of 7 to 21 years.
For the purpose of our study, we selectively include datasets sourced from distinct sites, namely the New York University medical center (NYU), Kennedy Krieger Institute (KKI), and Peking University (PU) sites. Each site's dataset harbors unique characteristics, which are succinctly outlined in Table 1.
Table 1. Details of ADHD-200 dataset
Site | Age | Female | Male | Healthy Control | ADHD | Total |
NYU | 7–18 | 77 | 145 | 99 | 123 | 222 |
KKI | 8–13 | 37 | 46 | 61 | 22 | 83 |
NI | 11–22 | 17 | 31 | 23 | 25 | 48 |
PU | 8–17 | 52 | 142 | 116 | 78 | 194 |
PU_1 | 8–17 | 36 | 49 | 62 | 24 | 86 |
To streamline data sharing and preprocessing procedures, initiatives such as the R-fMRI maps project were instrumental. The datasets, including both the hold-out testing dataset and a subset of the original training dataset, underwent rigorous preprocessing utilizing the Data Processing Assistant for Resting-State fMRI (DPARSF) programs.
The preprocessing pipeline encompassed several vital steps to ensure data quality and consistency. These procedures encompassed bandpass filtering to isolate pertinent frequency components, slice measuring modification, head movement correction, regression of nuisance covariates, spatial coregistration, bandpass filtering to isolate relevant frequency components, normalization to a standardized space for comparability, and finally, smoothing with a Gaussian kernel to enhance spatial coherence.
3.2. Result analysis and discussion
The experiments utilized Intel Xeon Platinum 8171M CPUs and Nvidia GeForce RTX 3090 GPUs, with the model implemented under the Keras framework.
Comprehensive comparisons were conducted with existing methods to evaluate the effectiveness of the proposed approach. Benchmarks included various machine learning techniques previously utilized in fMRI-based diagnosis, such as Fusion fMRI [12], L1BioSVM [13], and R-Relief [14]. Additionally, the performance of deep learning models including 3D CNN [15], Deep fMRI [17], KD-Transformer [18], and STAAE [19] were evaluated.
Results from the comparison indicated that the proposed model outperformed both traditional machine learning and state-of-the-art deep learning methods (see Table 2).
Table 2. Comparison with existing methods
NYU | KKI | NI | PU | PU_1 | |
Machine learning | |||||
Fusion fMRI (2018) | 52.7 | 86.7 | – | – | 85.8 |
L1BioSVM (2018) | – | 81.3 | – | 81.1 | 86.7 |
R-Relief (2019) | 70.7 | 81.8 | – | 68.6 | – |
Deep learning | |||||
3D CNN (2019) | 70.5 | 63.0 | – | 72.8 | – |
Deep fMRI (2020) | 73.1 | – | 67.9 | – | 62.7 |
KD-Transformer (2022) | 82.9 | 90.9 | 72.0 | 70.6 | – |
STAAE (2022) | 82.2 | 76.6 | 63.7 | 79.5 | – |
proposed method | |||||
CBAM-Autoencoder | 91.7 | 93.8 | 86.4 | 89.1 | 83.5 |
4. Conclusion
In summary, this study introduces an innovative method for accurately detecting Attention Deficit Hyperactivity Disorder (ADHD) using resting-state functional Magnetic Resonance Imaging (fMRI) data. The proposed approach combines the Convolutional Block Attention Module (CBAM) with a lightweight Autoencoder network to effectively extract and emphasize key features within fMRI scans. Through the integration of attention mechanisms, the model prioritizes crucial local details while minimizing irrelevant information, thus enhancing diagnostic accuracy.
Extensive experimentation on the ADHD-200 dataset demonstrates the effectiveness of the proposed method, resulting in significant improvements in classification performance. Specifically, the approach achieved notable average accuracies of 91.7% across the NYU, 93.8% across the KKI, 86.4% across the NI, 89.1% across the PU, and 83.5% across the PU_1 datasets.
The proposed CBAM-autoencoder network architecture offers several contributions. Firstly, it presents a novel approach that enhances classification accuracy on the ADHD-200 dataset. Secondly, by removing the decoder post-training and retaining only the encoder and classification layers for prediction, the framework optimizes model efficiency. Lastly, the integration of CBAM enhances the network's ability to concentrate on pertinent features while disregarding irrelevant ones.
Although the ADHD-200 dataset is the only one the study looks at, more research is needed to determine whether the suggested strategy can be executed in real-world healthcare environments and if it can be generalized to other datasets. This will help advance the field of ADHD diagnosis and treatment approaches.
Acknowledgement
This work was supported in part by the top-notched student program of Sichuan University.
References
[1]. American Psychiatric Association. (2013). Diagnostic and Statistical Manual of Mental Disorders: DSM-5 (5th ed.). Arlington, VA: American Psychiatric Association.
[2]. Tamam, L., Karakus, G., & Ozpoyraz, N. (2008). Comorbidity of adult attention-deficit hyperactivity disorder and bipolar disorder: Prevalence and clinical correlates. European Archives of Psychiatry and Clinical Neuroscience, 258, 385–393. https://doi.org/10.1007/s00406-008-0807-x.
[3]. Klassen, L. J., Katzman, M. A., & Chokka, P. (2009). Adult ADHD and its comorbidities, with a focus on bipolar disorder. Journal of Affective Disorders, 124(1–2), 1–8.
[4]. Conners, C. K., Pitkanen, J., & Rzepa, S. R. (2011). Conners (Conners 3; Conners 2008) (3rd ed.). New York, NY, USA: Springer. pp. 675-678.
[5]. Loh, H. W., Ooi, C. P., Barua, P. D., Palmer, E. E., Molinari, F., & Acharya, U. R. (2022). Automated detection of ADHD: Current trends and future perspective. Computers in Biology and Medicine, 146, 105525. doi: 10.1016/j.compbiomed.2022.105525.
[6]. Milham, M. P., Fair, D., Mennes, M., & Mostofsky, S. H. (2012). The ADHD-200 consortium: A model to advance the translational potential of neuroimaging in clinical neuroscience. Frontiers in Systems Neuroscience, 6, 62.
[7]. Huettel, S. A., Song, A. W., & McCarthy, G. (2009). Functional Magnetic Resonance Imaging. Yale Journal of Biology and Medicine, 82(4), 233.
[8]. Eslami, T., Almuqhim, F., Raiker, J. S., & Saeed, F. (2021). Machine learning methods for diagnosing autism spectrum disorder and attention-deficit/hyperactivity disorder using functional and structural MRI: A survey. Frontiers in Neuroinformatics, 14. doi: 10.3389/fninf.2020.623543.
[9]. Dey, S., Rao, A. R., & Shah, M. (2014). Attributed graph distance measure for automatic detection of attention deficit hyperactive disordered subjects. Frontiers in Neural Circuits, 8. https://doi.org/10.3389/fncir.2014.00064.
[10]. Colby, J., Rudie, J., Brown, J., Douglas, P., Cohen, M., & Shehzad, Z. (2012). Insights into multimodal imaging classification of ADHD. Frontiers in Systems Neuroscience, 6. https://doi.org/10.3389/fnsys.2012.00059
[11]. Riaz, A., Alonso, E., & Slabaugh, G. (2016). Phenotypic Integrated Framework for Classification of ADHD Using fMRI Image Analysis and Recognition. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Volume 9730 (pp. [page range]). ISBN: 978-3-319-41500-0.
[12]. Riaz, A., Asad, M., Alonso, E., & Slabaugh, G. (2018). Fusion of fMRI and non-imaging data for ADHD classification. Computerized Medical Imaging and Graphics, 65, 115–128.
[13]. Riaz, A., Asad, M., Alonso, E., & Slabaugh, G. (2018). Fusion of fMRI and non-imaging data for ADHD classification. Computerized Medical Imaging and Graphics, 65, 115–128.
[14]. Miao, B., Zhang, L. L., Guan, J. L., Meng, Q. F. M., & Zhang, Y. L. (2019). Classification of ADHD individuals and neurotypicals using reliable RELIEF: A resting-state study. IEEE Access, 7, 62163–62171.
[15]. Zou, L., Zheng, J., Miao, C., McKeown, M. J., & Wang, Z. J. (2017). 3D CNN Based Automatic Diagnosis of Attention Deficit Hyperactivity Disorder Using Functional and Structural MRI. IEEE Access, 5, 23626-23636. doi: 10.1109/ACCESS.2017.2762703.
[16]. Mao, Z., Su, Y., Xu, G., Wang, X., Huang, Y., Yue, W., Sun, L., & Xiong, N. (2019). Spatio-temporal deep learning method for ADHD fMRI classification. Information Sciences, 499, 1-11. https://doi.org/10.1016/j.ins.2019.05.043.
[17]. Riaz, A., Asad, M., Alonso, E., & Slabaugh, G. (2020). DeepFMRI: End-to-end deep learning for functional connectivity and classification of ADHD using fMRI. Journal of Neuroscience Methods, 335, 108506.
[18]. Zhang, J., Zhou, L., Wang, L., Liu, M., & Shen, D. (2022). Diffusion Kernel Attention Network for Brain Disorder Classification. IEEE Transactions on Medical Imaging, 41(10), 2814-2827. doi: 10.1109/TMI.2022.3170701.
[19]. Qiang, N., Dong, Q., Liang, H., et al. (2022). A novel ADHD classification method based on resting state temporal templates (RSTT) using spatiotemporal attention auto-encoder. Neural Computing & Applications, 34, 7815–7833. https://doi.org/10.1007/s00521-021-06868-w.
[20]. Woo, S., Park, J., Lee, J. Y., & Kweon, I. S. (2018). Cbam: Convolutional block attention module. In Proceedings of the European conference on computer vision (ECCV) (pp. 3-19).
Cite this article
Hong,S. (2024). ADHD fMRI classification through CBAM-Autoencoder framework. Applied and Computational Engineering,88,12-19.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 6th International Conference on Computing and Data Science
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. American Psychiatric Association. (2013). Diagnostic and Statistical Manual of Mental Disorders: DSM-5 (5th ed.). Arlington, VA: American Psychiatric Association.
[2]. Tamam, L., Karakus, G., & Ozpoyraz, N. (2008). Comorbidity of adult attention-deficit hyperactivity disorder and bipolar disorder: Prevalence and clinical correlates. European Archives of Psychiatry and Clinical Neuroscience, 258, 385–393. https://doi.org/10.1007/s00406-008-0807-x.
[3]. Klassen, L. J., Katzman, M. A., & Chokka, P. (2009). Adult ADHD and its comorbidities, with a focus on bipolar disorder. Journal of Affective Disorders, 124(1–2), 1–8.
[4]. Conners, C. K., Pitkanen, J., & Rzepa, S. R. (2011). Conners (Conners 3; Conners 2008) (3rd ed.). New York, NY, USA: Springer. pp. 675-678.
[5]. Loh, H. W., Ooi, C. P., Barua, P. D., Palmer, E. E., Molinari, F., & Acharya, U. R. (2022). Automated detection of ADHD: Current trends and future perspective. Computers in Biology and Medicine, 146, 105525. doi: 10.1016/j.compbiomed.2022.105525.
[6]. Milham, M. P., Fair, D., Mennes, M., & Mostofsky, S. H. (2012). The ADHD-200 consortium: A model to advance the translational potential of neuroimaging in clinical neuroscience. Frontiers in Systems Neuroscience, 6, 62.
[7]. Huettel, S. A., Song, A. W., & McCarthy, G. (2009). Functional Magnetic Resonance Imaging. Yale Journal of Biology and Medicine, 82(4), 233.
[8]. Eslami, T., Almuqhim, F., Raiker, J. S., & Saeed, F. (2021). Machine learning methods for diagnosing autism spectrum disorder and attention-deficit/hyperactivity disorder using functional and structural MRI: A survey. Frontiers in Neuroinformatics, 14. doi: 10.3389/fninf.2020.623543.
[9]. Dey, S., Rao, A. R., & Shah, M. (2014). Attributed graph distance measure for automatic detection of attention deficit hyperactive disordered subjects. Frontiers in Neural Circuits, 8. https://doi.org/10.3389/fncir.2014.00064.
[10]. Colby, J., Rudie, J., Brown, J., Douglas, P., Cohen, M., & Shehzad, Z. (2012). Insights into multimodal imaging classification of ADHD. Frontiers in Systems Neuroscience, 6. https://doi.org/10.3389/fnsys.2012.00059
[11]. Riaz, A., Alonso, E., & Slabaugh, G. (2016). Phenotypic Integrated Framework for Classification of ADHD Using fMRI Image Analysis and Recognition. In Proceedings of the International Conference on Medical Image Computing and Computer-Assisted Intervention (MICCAI), Volume 9730 (pp. [page range]). ISBN: 978-3-319-41500-0.
[12]. Riaz, A., Asad, M., Alonso, E., & Slabaugh, G. (2018). Fusion of fMRI and non-imaging data for ADHD classification. Computerized Medical Imaging and Graphics, 65, 115–128.
[13]. Riaz, A., Asad, M., Alonso, E., & Slabaugh, G. (2018). Fusion of fMRI and non-imaging data for ADHD classification. Computerized Medical Imaging and Graphics, 65, 115–128.
[14]. Miao, B., Zhang, L. L., Guan, J. L., Meng, Q. F. M., & Zhang, Y. L. (2019). Classification of ADHD individuals and neurotypicals using reliable RELIEF: A resting-state study. IEEE Access, 7, 62163–62171.
[15]. Zou, L., Zheng, J., Miao, C., McKeown, M. J., & Wang, Z. J. (2017). 3D CNN Based Automatic Diagnosis of Attention Deficit Hyperactivity Disorder Using Functional and Structural MRI. IEEE Access, 5, 23626-23636. doi: 10.1109/ACCESS.2017.2762703.
[16]. Mao, Z., Su, Y., Xu, G., Wang, X., Huang, Y., Yue, W., Sun, L., & Xiong, N. (2019). Spatio-temporal deep learning method for ADHD fMRI classification. Information Sciences, 499, 1-11. https://doi.org/10.1016/j.ins.2019.05.043.
[17]. Riaz, A., Asad, M., Alonso, E., & Slabaugh, G. (2020). DeepFMRI: End-to-end deep learning for functional connectivity and classification of ADHD using fMRI. Journal of Neuroscience Methods, 335, 108506.
[18]. Zhang, J., Zhou, L., Wang, L., Liu, M., & Shen, D. (2022). Diffusion Kernel Attention Network for Brain Disorder Classification. IEEE Transactions on Medical Imaging, 41(10), 2814-2827. doi: 10.1109/TMI.2022.3170701.
[19]. Qiang, N., Dong, Q., Liang, H., et al. (2022). A novel ADHD classification method based on resting state temporal templates (RSTT) using spatiotemporal attention auto-encoder. Neural Computing & Applications, 34, 7815–7833. https://doi.org/10.1007/s00521-021-06868-w.
[20]. Woo, S., Park, J., Lee, J. Y., & Kweon, I. S. (2018). Cbam: Convolutional block attention module. In Proceedings of the European conference on computer vision (ECCV) (pp. 3-19).









