1. Introduction
Skin disease image segmentation is a critical component of computer-aided diagnosis, providing precise localization and delineation of lesions, thereby improving diagnostic accuracy and efficiency. In recent years, with the development of deep learning technologies, especially advancements in convolutional neural networks (CNNs), significant breakthroughs have been achieved in the field of medical image segmentation. From early fully convolutional networks (FCNs) to subsequent U-Net and its variants [1,2], and even to complex networks utilizing attention mechanisms and Transformer structures [3,5], the performance of neural network models in image segmentation is continuously improving.
However, the segmentation performance of existing models in the field of skin disease image segmentation still has considerable room for improvement and potential for growth. This is primarily due to the following issues: first, the diversity and complexity of skin lesions, including irregular shapes, color variations, and background interference, make it challenging for models to distinguish between normal tissues and lesion areas [6-9]. Secondly, existing models often neglect the balance between local and global information [3], resulting in decreased segmentation accuracy when dealing with blurred edges and complex structures.
To overcome these challenges, we proposed DMDLK-Net, adopting a unique approach by introducing a dynamic decoding strategy combined with a multi-scale feature fusion, enhancing the ability to capture and refine skin lesion features. This design improves the sensitivity of the model in capturing fine structures and complex textures. In the experiments section, we will demonstrate the performance of DMDLK-Net on skin disease image segmentation datasets, proving its superiority in segmentation accuracy.
The main contributions of this paper are as follows:
1. We introduced deformable large kernels (DLK) to address the morphological variability of skin lesion regions, designing the DDLK module for flexible feature extraction.
2. We design a DMFF module to dynamically achieve multi-scale feature integration by channel and spatial attention enhancement with less information losing.
3. We present a network which connects features in different level. The model outperforms existing state-of-the-art models, significantly improving segmentation accuracy.
In the upcoming part, we first provide a detailed introduction to the network structure, particularly focusing on the two key components in Section 2. Experiments and analysis are drawn in Section 3. Finally, some conclusions are provided in Section4.
2. Methods
2.1. Dynamic Deformable Large Kernel (DDLK)
Based on the DLK module of D-Net [4], we improved it and proposed the DDLK module using deformable large kernels for dynamic feature extraction. The deformable large kernel (DLK) can flexibly distort receptive fields, allowing the model to adapt to different data patterns [6, 7, 8, 9]. For segmentation tasks of skin diseases with diverse shapes and variable morphology, such flexible kernel shapes can enhance the representation of lesion areas, improving the definition of object contours and feature extraction accuracy. The structure of the deformable large kernel is shown in Figure 1.
The DDLK module first uses two serially connected DLKs with different kernel sizes to capture both local feature \( {M_{1}} \) and global feature \( {M_{2}} \) :
\( {M_{1}}, {M_{2}}=DLKConv(G), DLKConv(DLKConv(G)) \)
where \( G \) is the input feature. The generated feature maps are concatenated, and then global average pooling and global max pooling are applied to aggregation features in the spatial dimension. Then we adopt a convolution layer for feature interaction and a Sigmoid activation for information selection, and dynamically enhance vital information, obtaining feature \( {M^{*}} \) :
\( {M_{p}}=Concat(AveP(Concat({M_{1}},{M_{2}})),MaxP(Concat({M_{1}},{M_{2}}))) \)
\( M_{1}^{*}={M_{1}}⊗Sigmoid({Conv_{1×1}}({M_{p}})) \)
\( M_{2}^{*}={M_{2}}⊗Sigmoid({Conv_{1×1}}({M_{p}})) \)
\( {M^{*}}=M_{1}^{*}⊕M_{2}^{*} \)
By adding residual connections with the input feature map, prominent information in the channel dimension of the lesion features can be better replenished and refined:
\( {G^{*}}=G⊕{M^{*}} \)
where " \( ⊗ \) " represents pixel-wise multiplication, and " \( ⊕ \) " represents pixel-wise addition.
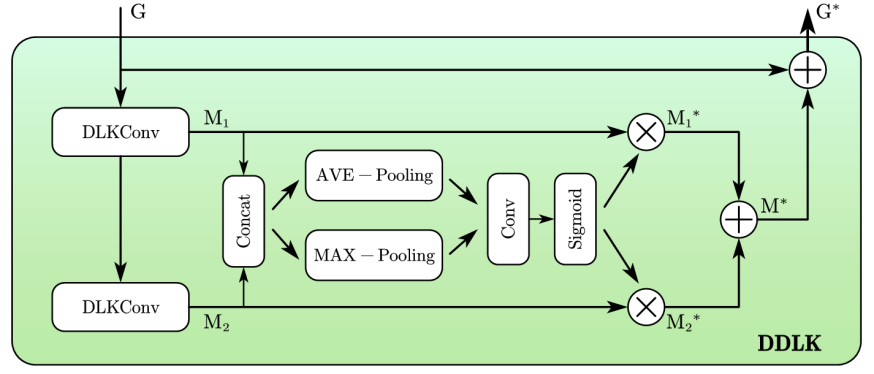
Figure 1. DDLK
2.2. Dynamic Multi-Scale Feature Fusion (DMFF)
Inspired by Jin Yang et al. [4] and Huajun Liu et al. [10], we propose a DMFF module. As shown in Figure 3, given two input features, \( {G_{1}} \) and \( {G_{2}} \) , which come from different layers of the model, the DMFF module selectively fuses different features in both spatial and channel dimensional enhancement .
In the spatial attention part, \( {G_{1}} \) and \( {G_{2}} \) are downsampled to the size of \( 1×H×W \) by using \( 1×1 \) convolutions, and then are fused to generate the integrated feature \( {M_{S}} \) :
\( {G_{1S}}={Conv_{1×1}}({G_{1}}) \)
\( {G_{2S}}={Conv_{1×1}}({G_{2}}) \)
\( {M_{S}}=Sigmoid({G_{1S}}⊕{G_{2S}}) \)
where " \( ⊕ \) " represents pixel-wise addition. In the channel attention part, \( {G_{1}} \) and \( {G_{2}} \) are firstly concatenated and then processed through three parallel paths. In the uppermost path, \( {G_{C}} \) is downsampled in channel dimension to \( 1×H×W \) through a \( 1×1 \) convolution, reshaped to \( HW×1 \) , and then passed through a Softmax layer to highlight the channel features. In the other parallel path, \( {G_{C}} \) is downsampled to \( C/2×H×W \) through a \( 1×1 \) convolution and reshaped to \( C/2×HW \) for subsequent feature refinement and then obtain feature \( {G_{C-M}} \) , which integrates spatial and channel information and enhance the feature contrast in channel dimension.
\( {G_{C-U}}=Softmax({Reshape_{HW×1}}({Conv_{1×1}}(Concat({G_{1}},{G_{2}})))) \)
\( {G_{C-M}}={Reshape_{C/2×HW}}({Conv_{1×1}}({G_{C}})) \)
\( {G_{C-UM}}={G_{C-M}}×{G_{C-U}} \)
where " \( × \) " represents matrix multiplication. After additionally processed through a \( 1×1 \) convolution, a layer normalization, and a Sigmoid function, the activated feature integrates with the residual connection of \( {G_{C}} \) , resulting in the enriched feature \( {M_{C}} : \)
\( G_{C}^{*}={G_{C}}⊗Sigmoid(LN({Conv_{1×1}}({G_{C-UM}}))) \)
\( {M_{C}}=({Conv_{1×1}}(G_{C}^{*})) \)
Finally, the features dynamically selected by spatial and channel information are fused into \( {G^{*}} \) to complete selected feature fusion: \( { G^{*}}={M_{s}}⊗{M_{C}} \) .
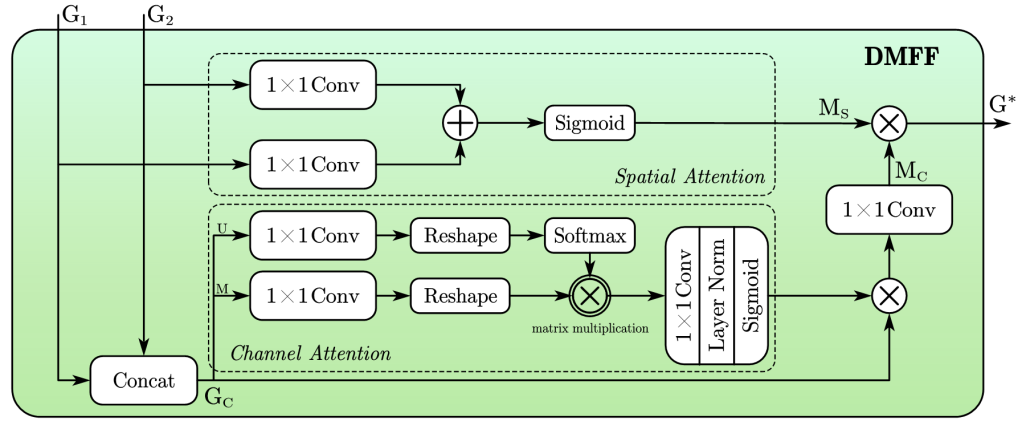
Figure 2. DMFF
2.3. DMDLK-Net Architecture
The architecture of DMDLK-Net is illustrated in Figure 3. It follows a classic symmetric encoder-decoder architecture and is organized into four levels.
In the encoder, we use the Mobile Convolution and Attention (MOAT) module for encoding tasks [5]. The structure of the MOAT module is shown on the left side of Figure 3. It sequentially consists of a lightweight convolutional part with depthwise separable convolutions for feature extraction and a self-attention block for attention enhancement, maintaining high feature extraction and expression capabilities while having a simple structure and low computational complexity.
The encoder is divided into four levels, with each level downsampling the input image by half. The first level is the STEM layer, composed of simple convolutions. The second to fourth levels consist of dual, triple, and heptuple stacked MOAT modules, respectively. In each stack of MOATs, the top MOAT module is responsible for downsampling, while the remaining modules enhance feature extraction. After downsampling the input image to 1/16th of its size, it is fed into the decoder through a symmetric bottleneck structure with down-up sampling.
The decoder is also divided into four levels, corresponding one-to-one with the encoder levels. One of our major innovations is incorporating three levels of skip connections (Upsampling skip, Downsampling skip, Identity skip) at each level of the decoder, passing the feature from the lower, corresponding, and upper levels of the encoder to the decoder, respectively, for feature concatenation. This allows the capture of both more global (lower level) and finer (upper level) abstract information.
During the bottom-up process, the DMFF module dynamically fuses the concatenated features from the encoder with the upsampled features from the deeper decoder levels. The resulting feature is passed to the DDLK module for further dynamic feature extraction and attention enhancement, finally being passed to the deeper levels of the decoder. At each level, the feature map is upsampled to twice its size, and ultimately, the final output is produced at the input size.
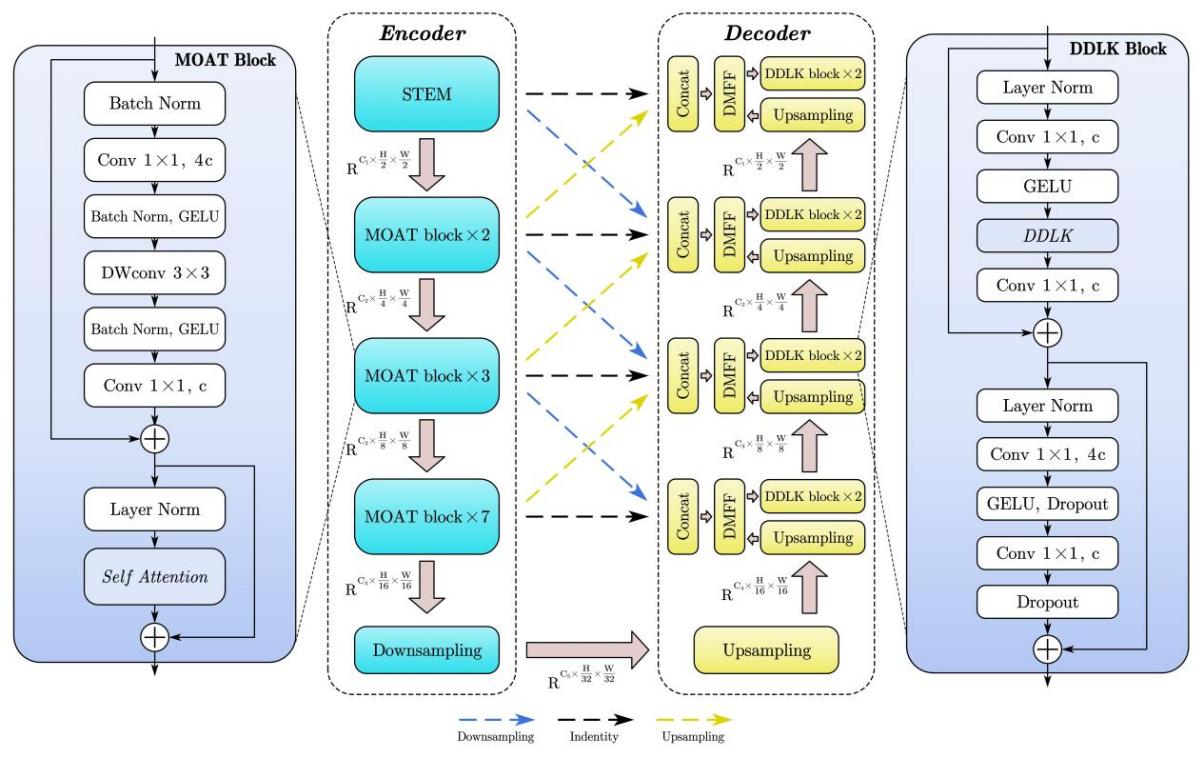
Figure 3. Overall Architecture
3. Experiment
3.1. Datasets
Our experiments primarily utilize the publicly available ISIC (International Skin Imaging Collaboration) 2018 dataset. The ISIC 2018 dataset is widely used for skin lesion medical image analysis. The segmentation task contains 2,594 images for the training set, 100 images for the validation set, and 1,000 images for the test set, with lesion labels annotated by experts. To enrich data and enhance the robustness of the model, we performed data augmentation: each image was randomly subjected to horizontal flipping, vertical flipping, rotation, scaling, translation, color jittering, and advanced blurring, expanding the training set to three times its original size, resulting in nearly 7,800 images. All input images were resized to a dimension of \( 448×448 \) .
3.2. Details
We use the cross-entropy function as the loss function, adopt the Adam algorithm as the optimizer, set the learning rate to \( 0.0001 \) , batch size to \( 4 \) , and trained for 200 epochs. The model training was conducted on a GPU platform with NVIDIA RTX 3090s.
3.3. Results
We tested the trained model on the test set containing 1,000 images to evaluate the model's performance and compared it with existing models ,as shown in Table 1. To access the accuracy of the segmentation results, we used Pixel Accuracy(Acc), Intersection over Union(IoU), and Dice Coefficient(Dice) as evaluation metrics.
Table 1. Comparison of eleven existing models
IoU | Dice | Acc | |
UNet | 78.14 | 86.28 | 91.92 |
UNet++ | 79.61 | 87.27 | 92.13 |
Attention-UNet | 79.61 | 87.42 | 92.74 |
UTNet | 78.50 | 86.29 | 92.16 |
SegFormer | 79.96 | 87.48 | 92.72 |
Swin-Unet | 79.74 | 87.02 | 92.19 |
UNext | 77.36 | 85.63 | 91.75 |
MALUNet | 79.73 | 87.59 | 92.53 |
UCTransNet | 79.67 | 87.19 | 92.46 |
DCSAU-Net | 79.00 | 86.80 | 92.12 |
EGE-UNet | 78.45 | 86.12 | 92.05 |
Average | 79.07 | 86.83 | 92.25 |
DMDLK-Net | 79.65 | 87.05 | 94.44 |
Our DMDLK-Net achieved the best performance in the Acc, significantly surpassing the other eleven models in the comparison experiments. Our model is 1.70% higher than the best-perform model in Acc, with IoU and Dice coefficients 0.58% and 0.22% higher than the average performance of these existing models, respectively.
Additionally, to visually demonstrate the effectiveness of our model, Figure 4 shows several representative segmentation examples on the ISIC-2018 test set, including the original image, ground truth, and the segmentation results of DMDLK-Net. Our model accurately delineates the lesion boundaries and details, highlighting its practical value in clinical applications.
The results demonstrate that our method has advantages in handling various skin disease lesion features and complex texture information. The introduction of deformable large kernels in the DDLK module enables more flexible and effective extraction of lesion features, significantly enhancing the accuracy of contour localization and edge delineation. The DMFF module, through multi-scale attention mechanisms, effectively integrates different levels of features, improving the representation capabilities and generalization performance of the model.
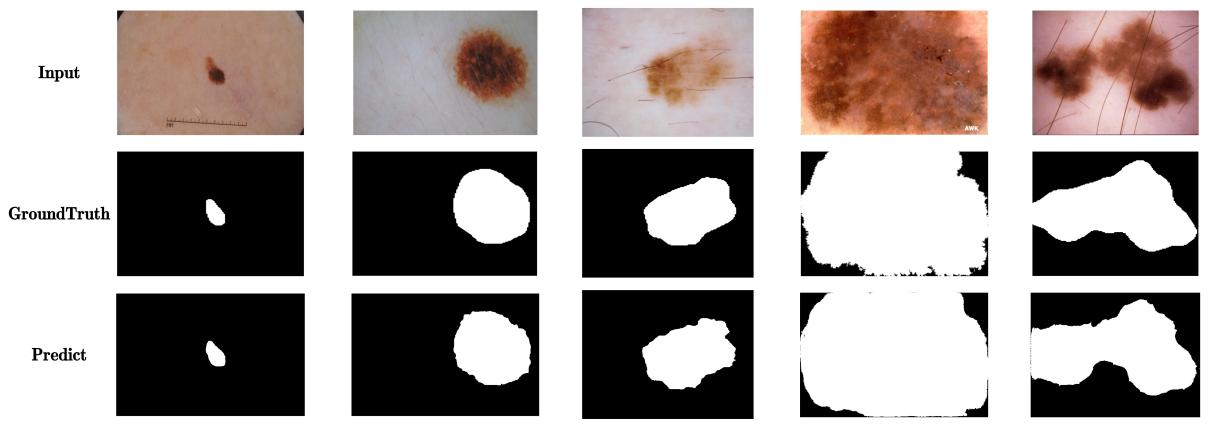
Figure 4. Visual display of the segmentation effect of DMDLK-Net
4. Conclusion
In this paper, we introduced DMDLK-Net, a novel network architecture designed to improve the segmentation of skin disease images by addressing the inherent challenges posed by the diversity and complexity of skin lesions. Our approach leverages the Dynamic Deformable Large Kernel (DDLK) module for flexible and accurate feature extraction and the Dynamic Multi-Scale Feature Fusion (DMFF) module for effective integration of multi-scale information. The comprehensive experiments conducted on the ISIC-2018 dataset demonstrate that DMDLK-Net outperforms existing models in Table 1. These results highlight the potential of DMDLK-Net to enhance clinical diagnostics by providing precise lesion segmentation. Future work will explore the extension of our network to other types of medical image segmentation tasks and further optimize the model for real-time applications. Additionally, integrating more advanced attention mechanisms and exploring the benefits of self-supervised learning could further enhance the performance and generalization capabilities.
References
[1]. Jonathan Long, Evan Shelhamer, Trevor Darrell. (2011). Fully Convolutional Networks for Semantic Segmentation. In: Computer Vision and Pattern Recognition. arXiv:1411.4038.
[2]. Olaf Ronneberger, Philipp Fischer, and Thomas Brox. (2015). U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Computer Vision and Pattern Recognition. arXiv: 1505.04597.
[3]. Alexey Dosovitskiy, Lucas Beyer, Alexander Kolesnikov, Dirk Weissenborn, Xiaohua Zhai, Thomas Unterthiner, Mostafa Dehghani, Matthias Minderer, Georg Heigold, Sylvain Gelly, Jakob Uszkoreit, Neil Houlsby. (2020). An Image Is Worth 16x16 Words: Transformers For Image Recognition At Scale. In: Computer Vision and Pattern Recognition. arXiv:2010.11929.
[4]. Jin Yang, Peijie Qiu, Yichi Zhang, Daniel S. Marcus, Aristeidis Sotiras. (2024). D-Net: Dynamic Large Kernel with Dynamic Feature Fusion for Volumetric Medical Image Segmentation. In: Computer Vision and Pattern Recognition. arXiv:2403.10674.
[5]. Chenglin Yang, Siyuan Qiao, Qihang Yu, Xiaoding Yuan, Yukun Zhu, Alan Yuille, Hartwig Adam, Liang-Chieh Chen. (2022). MOAT: Alternating Mobile Convolution and Attention Brings Strong Vision Models. In: Computer Vision and Pattern Recognition. arXiv:2210.01820.
[6]. Reza Azad, Leon Niggemeier, Michael Hüttemann, Amirhossein Kazerouni, Ehsan Khodapanah Aghdam, Yury Velichko, Ulas Bagci, Dorit Merhof. (2023). Beyond Self Attention: Deformable Large Kernel Attention for Medical Image Segmatation. In: Computer Vision and Pattern Recognition. arXiv:2309.00121.
[7]. Ding, X., Zhang, X., Han, J., Ding, G. (2023). Scaling up your kernels to 31x31: Revisiting large kernel design in cnns. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition. pp. 11963–11975. arXiv:2303.09030.
[8]. Li, Y., Hou, Q., Zheng, Z., Cheng, M.M., Yang, J., Li, X. (2023). Large selective kernel network for remote sensing object detection. arXiv:2303.09030
[9]. Liu, Z., Mao, H., Wu, C.Y., Feichtenhofer, C., Darrell, T., Xie, S. (2022). A convnet for the 2020s. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition. pp. 11976–11986. arXiv:2201.03545.
[10]. Huajun Liu, Fuqiang Liu, Xinyi Fan, Dong Huang. (2021) Polarized Self-Attention: Towards High-quality Pixel-wise Regression. In: Computer Vision and Pattern Recognition. arXiv: 2107.00782.
Cite this article
Wei,B. (2024). DMDLK-Net: A dynamic multi-scale feature fusion network with deformable large kernel for medical segmentation. Applied and Computational Engineering,95,79-85.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 6th International Conference on Computing and Data Science
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Jonathan Long, Evan Shelhamer, Trevor Darrell. (2011). Fully Convolutional Networks for Semantic Segmentation. In: Computer Vision and Pattern Recognition. arXiv:1411.4038.
[2]. Olaf Ronneberger, Philipp Fischer, and Thomas Brox. (2015). U-Net: Convolutional Networks for Biomedical Image Segmentation. In: Computer Vision and Pattern Recognition. arXiv: 1505.04597.
[3]. Alexey Dosovitskiy, Lucas Beyer, Alexander Kolesnikov, Dirk Weissenborn, Xiaohua Zhai, Thomas Unterthiner, Mostafa Dehghani, Matthias Minderer, Georg Heigold, Sylvain Gelly, Jakob Uszkoreit, Neil Houlsby. (2020). An Image Is Worth 16x16 Words: Transformers For Image Recognition At Scale. In: Computer Vision and Pattern Recognition. arXiv:2010.11929.
[4]. Jin Yang, Peijie Qiu, Yichi Zhang, Daniel S. Marcus, Aristeidis Sotiras. (2024). D-Net: Dynamic Large Kernel with Dynamic Feature Fusion for Volumetric Medical Image Segmentation. In: Computer Vision and Pattern Recognition. arXiv:2403.10674.
[5]. Chenglin Yang, Siyuan Qiao, Qihang Yu, Xiaoding Yuan, Yukun Zhu, Alan Yuille, Hartwig Adam, Liang-Chieh Chen. (2022). MOAT: Alternating Mobile Convolution and Attention Brings Strong Vision Models. In: Computer Vision and Pattern Recognition. arXiv:2210.01820.
[6]. Reza Azad, Leon Niggemeier, Michael Hüttemann, Amirhossein Kazerouni, Ehsan Khodapanah Aghdam, Yury Velichko, Ulas Bagci, Dorit Merhof. (2023). Beyond Self Attention: Deformable Large Kernel Attention for Medical Image Segmatation. In: Computer Vision and Pattern Recognition. arXiv:2309.00121.
[7]. Ding, X., Zhang, X., Han, J., Ding, G. (2023). Scaling up your kernels to 31x31: Revisiting large kernel design in cnns. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition. pp. 11963–11975. arXiv:2303.09030.
[8]. Li, Y., Hou, Q., Zheng, Z., Cheng, M.M., Yang, J., Li, X. (2023). Large selective kernel network for remote sensing object detection. arXiv:2303.09030
[9]. Liu, Z., Mao, H., Wu, C.Y., Feichtenhofer, C., Darrell, T., Xie, S. (2022). A convnet for the 2020s. In: Proceedings of the IEEE/CVF conference on computer vision and pattern recognition. pp. 11976–11986. arXiv:2201.03545.
[10]. Huajun Liu, Fuqiang Liu, Xinyi Fan, Dong Huang. (2021) Polarized Self-Attention: Towards High-quality Pixel-wise Regression. In: Computer Vision and Pattern Recognition. arXiv: 2107.00782.









