1. Introduction
Cancer remains one of the most lethal diseases in modern society, and its global impact is staggering. According to statistical projections, the global burden of cancer is expected to reach 28.4 million cases by 2040, representing a significant challenge for healthcare systems worldwide [1]. The rising incidence of cancer underscores the urgent need for advancements in early diagnosis and treatment, which can drastically improve patient outcomes. In this context, the advent of computer-aided diagnosis (CAD) has introduced a transformative approach to medical image analysis, offering the potential to revolutionize clinical medicine by enhancing diagnostic accuracy, speed, and consistency [2].
Traditional image processing techniques, while valuable, often struggle to keep pace with the growing complexity and scale of modern medical data. Large-scale cancer image datasets, characterized by high dimensionality, heterogeneity, and subtle variations, present significant challenges. These methods frequently falter when confronted with such data, as they may be ill-equipped to effectively handle the intricate patterns and relationships within the images. To address these limitations, deep learning techniques, particularly convolutional neural networks (CNNs), have emerged as powerful tools for medical image analysis. CNNs have become integral to tasks such as image classification, target detection, and feature extraction, achieving unprecedented levels of performance in complex imaging tasks [3].
CNN architectures like VGGNet, ResNet, and Fully Convolutional Networks (FCN) have proven to be highly effective in medical image classification. These models leverage deep hierarchical structures to automatically learn relevant features from raw data, eliminating the need for manual feature engineering. Their ability to detect subtle patterns and abnormalities in medical images has positioned CNNs as essential components in cancer detection and diagnosis, facilitating earlier and more accurate identification of malignant tissues. Moreover, the inherent subjectivity in medical image interpretation poses additional challenges. The variability in diagnoses across different medical professionals can lead to inconsistent annotations, which directly impacts the performance of CNN models. This variability necessitates further research into techniques that can standardize annotations and ensure greater consistency across datasets. Techniques such as ensemble learning and model fusion may help mitigate the impact of these inconsistencies by combining predictions from multiple models to improve accuracy and robustness. Additionally, the integration of multimodal data—which combines information from multiple imaging sources, such as MRI, CT, and PET—can enhance CNN performance by leveraging the complementary strengths of each modality. This fusion of diverse data streams has the potential to significantly improve the accuracy of cancer detection, particularly in complex cases where a single modality may be insufficient.
This review explores the utilization of CNNs in the classification of cancer images, with the objective of providing a comprehensive overview of recent advancements and challenges in this field. By analyzing the extant research outcomes, this paper seeks to elucidate the considerable potential of CNN algorithms in the domain of cancer image analysis, which can facilitate more reliable data analysis support for clinical decision-making.
2. Literature Survey
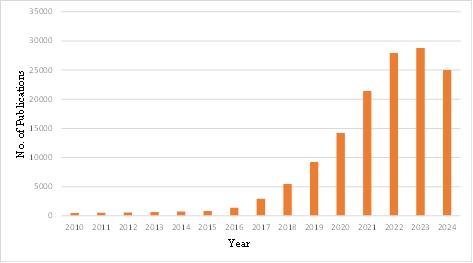
Figure 1. The number of papers searched using “CNN” and “Cancer Images Classification” per year
Figure 1 illustrates the annual number of publications retrieved using the keywords "CNN" and "cancer images classification" on Google Scholar. The number of relevant papers exhibited a gradual increase from 2010, with a pronounced surge in 2020, when the number of publications reached approximately 20,000. This upward trend continued, reaching a peak of 36,100 papers in 2023. The considerable increase in publications in 2020 can be attributed to the rapid advancement of AI technologies and the accessibility of a greater number of publicly available medical imaging datasets. These developments enabled researchers to train CNN models with more comprehensive data, thereby enhancing the accuracy and efficiency of cancer image classification. Overall, the number of publications has demonstrated a distinct upward trend, indicating a growing interest in the utilization of CNN models in cancer image classification. Furthermore, the total number of documents in 2024 is not yet fully determinable, resulting in an observable decline.
3. Application of CNNs in Cancer Image Classification
3.1. Types of Cancer Images
In the field of cancer research, various types of imaging data are crucial for diagnosis, treatment planning, and monitoring. The primary categories include histopathological images, radiological images, and molecular imaging, each offering unique insights into tumor characteristics and progression.
3.1.1. Histopathology Images
Histopathology images, also referred to as whole slide images (WSI) [4], are digital pathology images obtained using high-resolution scanning technology. They are typically stored in an image pyramid structure and have a resolution of up to 100,000 x 100,000 pixels. Furthermore, WSI offers numerous benefits. WSI provides high-quality and high-resolution images with annotations, reducing the risk of damage and fading during slide transport [5]. Additionally, WSI can display detailed information about tissue, cells, and subcellular structures, providing a reliable basis for digital pathological diagnosis. WSI pathological images remain consistent over time, and experts in different fields can collaborate through remote pathological networking systems to enhance the efficiency and accuracy of pathological diagnosis [6]. Moreover, WSI fosters collaboration among experts across different geographical locations through remote pathological networking systems. This capability not only enhances the efficiency of pathological diagnosis but also broadens the expertise available for complex cases, ultimately improving patient outcomes.
3.1.2. Radiological Images
Radiological images are pivotal in cancer detection and management, providing critical information in a non-invasive manner. Techniques such as computed tomography (CT) scans utilize X-rays to produce detailed cross-sectional images of the body, making them particularly effective for identifying tumors in internal organs, including those associated with lung and liver cancer. The speed and accessibility of CT scans make them a preferred choice for initial screenings and follow-up evaluations. In contrast, magnetic resonance imaging (MRI) employs strong magnetic fields and radio waves to generate high-resolution images of soft tissues. This modality is especially valuable for detecting cancers in areas such as the brain and breast, where the differentiation of soft tissue structures is crucial for accurate diagnosis and treatment planning. The detailed imaging capabilities of MRI allow for better visualization of tumor boundaries and involvement with surrounding tissues, guiding surgical and therapeutic interventions [7].
3.1.3. Molecular imaging
Molecular imaging represents a cutting-edge approach in cancer diagnostics, enabling the visualization of biological processes at the molecular and cellular levels. This technique helps identify metabolic irregularities, such as altered glucose metabolism, prior to the formation of tumor structures, which is critical for early detection. By providing detailed information regarding the location, size, and functionality of tumors, molecular imaging facilitates timely intervention and personalized treatment strategies [8]. Technologies such as positron emission tomography (PET) and single-photon emission computed tomography (SPECT) are commonly used in molecular imaging. These methods allow for the assessment of tumor biology and response to therapy, aiding in the evaluation of treatment efficacy over time. As a result, molecular imaging not only enhances diagnostic capabilities but also contributes to the ongoing development of targeted therapies and precision medicine in oncology.
3.2. CNN Models and Techniques
The field of medical image analysis is a significant area of study, with a particular focus on the detection and classification of cancer pathology images. The objective is to detect and identify the presence of cancer cells or tissues in pathological image data. While the utilization of deep learning for pathological image classification encompasses a multitude of disease categories, the predominant research methodologies are analogous and are founded upon convolutional neural network (CNN) architectures for classification. For example, Litjens et al. proposed a convolutional neural network (CNN)-based method for the detection of prostate cancer, which involved the segmentation of the slice image into small blocks and the subsequent extraction and classification of features using a CNN [9]. In their study, Varma et al. demonstrated that different convolutional neural network (CNN) models, including DenseNet-161 and ResNet-101, can automatically classify X-ray abnormalities of the foot, knee, ankle, and hip. Furthermore, they showed that CNNs achieve high area under the receiver operating characteristic curve (AUC-ROC) in this abnormal classification task [10].
The following section provides an overview of three of the most utilized convolutional neural network (CNN) structural models:
DenseNet-201: DenseNet-201 is a deep neural network architecture that employs dense connections. In the context of cancer pathology image classification, the DenseNet-201 model can be employed to extract high-level feature representations of the image, including local details and texture information, with the objective of enhancing the classification capacity of the classifier. Furthermore, the DenseNet model comprises DenseBlocks and an intermediate spacer module, Transition Layer. This model features a deeper network structure, enabling coverage of a broader range of scales and semantic information, and exhibits enhanced feature expression ability [11].
NasNetMobile: NasNetMobile, developed by the Google Brain team, is a lightweight deep neural network architecture designed to be computationally efficient with fewer parameters. This model is particularly well-suited for extracting features from pathological images on a smaller scale, allowing for effective analysis of cell morphology and tissue structures. Its reduced complexity enables rapid processing and deployment in clinical settings, making it a valuable resource for real-time cancer detection and classification [12].
VGG16: VGG16 is a well-established CNN architecture that has been pre-trained on the ImageNet dataset, frequently used in image classification and feature extraction tasks. In the context of pathological image classification, VGG16 effectively captures a broad spectrum of image features, ranging from low-level color and texture details to high-level semantic information. This model enhances the classifier's performance by leveraging its robust feature extraction capabilities, making it a reliable choice for analyzing cancer pathology images [13].
3.3. Transfer Learning in Cancer Pathology Image Analysis
The acquisition of high-quality medical image data and the labor-intensive, costly process of annotating such data pose significant challenges in the field of cancer pathology image analysis. These limitations have led to the widespread adoption of transfer learning techniques, wherein deep learning models pre-trained on large-scale datasets are adapted for medical image classification tasks. This approach leverages the learned features from large datasets such as ImageNet and applies them to specific domains like cancer detection. Pre-trained models have consistently been shown to function as highly effective feature extractors, often outperforming models trained from scratch on smaller, domain-specific datasets. Moreover, transfer learning accelerates model convergence, significantly reducing the time required to fine-tune models for high performance in medical applications [14, 15].
For instance, Xu et al. [16] proposed a lung cancer detection method that utilizes a pre-trained CNN model on the ImageNet dataset. In this approach, the pre-trained model serves as a feature extractor, and fine-tuning is performed using lung cancer tissue slice images. This strategy enhances the model's ability to recognize cancer-specific features while benefiting from the general knowledge learned from large, diverse datasets, leading to improved classification accuracy for lung cancer detection. In environments with limited computational resources, optimizing the performance of lightweight CNNs becomes crucial. Lightweight models offer reduced computational complexity and faster processing times, making them suitable for real-time applications in clinical settings. Studies have shown that lightweight CNN architectures can still achieve high levels of accuracy when effectively utilized. For example, Nobashi et al. [17] demonstrated that combining the strengths of multiple CNN models could improve performance in cancer image classification. Their study involved integrating various lightweight CNNs and comparing the performance of individual models against their integrated counterparts. The results indicated that the integrated models outperformed individual models, showcasing the potential of model fusion techniques to enhance classification performance, particularly in resource-constrained environments. Furthermore, Hung et al. [18] developed an automated method based on multimodal convolutional neural networks (CNNs) for distinguishing cancerous from non-cancerous tissues. Their approach integrated data from multiple imaging modalities, allowing the model to capture more comprehensive and complementary information for cancer diagnosis. By leveraging multimodal data, this method achieved improved accuracy in tissue classification, illustrating the advantages of integrating different types of data and CNN models to address the complexities of cancer pathology.
In addition to multimodal integration, other transfer learning strategies include domain adaptation and fine-tuning across different types of cancer pathology datasets. For example, pre-trained CNNs are often used as a base model, and only the final classification layers are retrained using task-specific data, such as breast or colorectal cancer images. This method allows the model to adapt quickly to new tasks while retaining the generalized feature extraction capabilities developed on larger datasets. Moreover, self-supervised learning and semi-supervised learning approaches are gaining traction as ways to overcome the data scarcity problem in medical imaging. These methods reduce reliance on large labeled datasets by leveraging unlabeled data for pre-training or by creating synthetic labels through techniques like data augmentation. Such strategies, combined with transfer learning, enable models to perform effectively even when labeled cancer pathology images are scarce.
4. Future Directions
From the reviewed papers, it is found that there has been a tremendous stride in analyzing medical images using CNN architectures. The utilization of lightweight architectural techniques serves not only to reduce the computational resource requirements of CNN models, but also to enhance the model performance on the original basis. Concurrently, the fusion of multimodal data has emerged as a pivotal element in enhancing diagnostic precision. The integration of multiple medical image data, including CT, MRI, and PET, enables CNN models to leverage the complementary characteristics of different image data, thereby enhancing the classification ability for complex pathologies. Furthermore, transfer learning techniques demonstrate considerable promise in the context of cancer image classification. By transferring the knowledge acquired by the model in one task to other tasks, particularly in the context of limited medical data, migration learning facilitates enhanced generalization and performance of the model. In the future, researchers may enhance model robustness through data augmentation techniques, adversarial training, and migration learning techniques, and improve model interpretability by combining visualization techniques such as Grad-CAM and models based on attention mechanisms.
5. Conclusion
Convolutional Neural Networks (CNNs) occupy a significant position in the domain of cancer image classification, as they markedly enhance the accuracy and efficiency of diagnosis in comparison to traditional methods. CNN models have exhibited exemplary performance in processing high-dimensional and intricate medical image data, particularly in the integration of multimodal image data, and have demonstrated remarkable adaptability. Nevertheless, significant challenges remain, including data scarcity, model interpretability, and large-scale clinical validation. To fully realize the potential of CNN technology, future research should prioritize the fusion of multimodal data, the development of interpretable techniques, and the application of lightweight architectures to enhance diagnostic efficacy and ensure the feasibility of practical clinical application.
References
[1]. Chhikara, B. S., & Parang, K. (2023). Global Cancer Statistics 2022: the trends projection analysis. Chemical Biology Letters, 10(1), 451-451.
[2]. Gao, Y., Geras, K. J., Lewin, A. A., & Moy, L. (2019). New frontiers: an update on computer-aided diagnosis for breast imaging in the age of artificial intelligence. American Journal of Roentgenology, 212(2), 300-307.
[3]. Cai, L., Gao, J., & Zhao, D. (2020). A review of the application of deep learning in medical image classification and segmentation. Annals of translational medicine, 8(11)
[4]. Dimitriou, N., Arandjelović, O., & Caie, P. D. (2019). Deep learning for whole slide image analysis: an overview. Frontiers in medicine, 6, 264.
[5]. Hanna, M. G., Parwani, A., & Sirintrapun, S. J. (2020). Whole slide imaging: technology and applications. Advances in Anatomic Pathology, 27(4), 251-259.
[6]. Li, X., Li, C., Rahaman, M. M., Sun, H., Li, X., Wu, J., ... & Grzegorzek, M. (2022). A comprehensive review of computer-aided whole-slide image analysis: from datasets to feature extraction, segmentation, classification and detection approaches. Artificial Intelligence Review, 55(6), 4809-4878.
[7]. Murtaza, G., Shuib, L., Abdul Wahab, A. W., Mujtaba, G., Mujtaba, G., Nweke, H. F., ... & Azmi, N. A. (2020). Deep learning-based breast cancer classification through medical imaging modalities: state of the art and research challenges. Artificial Intelligence Review, 53, 1655-1720.
[8]. Jiang, X., Hu, Z., Wang, S., & Zhang, Y. (2023). Deep learning for medical image-based cancer diagnosis. Cancers, 15(14), 3608.
[9]. Litjens, G., Kooi, T., Bejnordi, B. E., Setio, A. A. A., Ciompi, F., Ghafoorian, M., ... & Sánchez, C. I. (2017). A survey on deep learning in medical image analysis. Medical image analysis, 42, 60-88.
[10]. Varma, M., Lu, M., Gardner, R., Dunnmon, J., Khandwala, N., Rajpurkar, P., ... & Patel, B. N. (2019). Automated abnormality detection in lower extremity radiographs using deep learning. Nature Machine Intelligence, 1(12), 578-583.
[11]. Wang, S. H., & Zhang, Y. D. (2020). DenseNet-201-based deep neural network with composite learning factor and precomputation for multiple sclerosis classification. ACM Transactions on Multimedia Computing, Communications, and Applications (TOMM), 16(2s), 1-19.
[12]. Naskinova, I. (2023). Transfer learning with NASNet-Mobile for Pneumonia X-ray classification. Asian-European Journal of Mathematics, 16(01), 2250240.
[13]. Albashish, D., Al-Sayyed, R., Abdullah, A., Ryalat, M. H., & Almansour, N. A. (2021, July). Deep CNN model based on VGG16 for breast cancer classification. In 2021 International conference on information technology (ICIT) (pp. 805-810). IEEE.
[14]. Behar, N., & Shrivastava, M. (2022). ResNet50-Based Effective Model for Breast Cancer Classification Using Histopathology Images. CMES-Computer Modeling in Engineering & Sciences, 130(2).
[15]. Kora, P., Ooi, C. P., Faust, O., Raghavendra, U., Gudigar, A., Chan, W. Y., ... & Acharya, U. R. (2022). Transfer learning techniques for medical image analysis: A review. Biocybernetics and Biomedical Engineering, 42(1), 79-107.
[16]. Xu, H., Park, S., Lee, S. H., & Hwang, T. H. (2019). Using transfer learning on whole slide images to predict tumor mutational burden in bladder cancer patients. BioRxiv, 554527.
[17]. Nobashi, T., Zacharias, C., Ellis, J. K., Ferri, V., Koran, M. E., Franc, B. L., ... & Davidzon, G. A. (2020). Performance comparison of individual and ensemble CNN models for the classification of brain 18F-FDG-PET scans. Journal of Digital Imaging, 33, 447-455.
[18]. Le, M. H., Chen, J., Wang, L., Wang, Z., Liu, W., Cheng, K. T. T., & Yang, X. (2017). Automated diagnosis of prostate cancer in multi-parametric MRI based on multimodal convolutional neural networks. Physics in Medicine & Biology, 62(16), 6497.
Cite this article
Yi,H. (2024). A Review of Convolutional Neural Networks in Cancer Image Classification. Applied and Computational Engineering,97,69-74.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 2nd International Conference on Machine Learning and Automation
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Chhikara, B. S., & Parang, K. (2023). Global Cancer Statistics 2022: the trends projection analysis. Chemical Biology Letters, 10(1), 451-451.
[2]. Gao, Y., Geras, K. J., Lewin, A. A., & Moy, L. (2019). New frontiers: an update on computer-aided diagnosis for breast imaging in the age of artificial intelligence. American Journal of Roentgenology, 212(2), 300-307.
[3]. Cai, L., Gao, J., & Zhao, D. (2020). A review of the application of deep learning in medical image classification and segmentation. Annals of translational medicine, 8(11)
[4]. Dimitriou, N., Arandjelović, O., & Caie, P. D. (2019). Deep learning for whole slide image analysis: an overview. Frontiers in medicine, 6, 264.
[5]. Hanna, M. G., Parwani, A., & Sirintrapun, S. J. (2020). Whole slide imaging: technology and applications. Advances in Anatomic Pathology, 27(4), 251-259.
[6]. Li, X., Li, C., Rahaman, M. M., Sun, H., Li, X., Wu, J., ... & Grzegorzek, M. (2022). A comprehensive review of computer-aided whole-slide image analysis: from datasets to feature extraction, segmentation, classification and detection approaches. Artificial Intelligence Review, 55(6), 4809-4878.
[7]. Murtaza, G., Shuib, L., Abdul Wahab, A. W., Mujtaba, G., Mujtaba, G., Nweke, H. F., ... & Azmi, N. A. (2020). Deep learning-based breast cancer classification through medical imaging modalities: state of the art and research challenges. Artificial Intelligence Review, 53, 1655-1720.
[8]. Jiang, X., Hu, Z., Wang, S., & Zhang, Y. (2023). Deep learning for medical image-based cancer diagnosis. Cancers, 15(14), 3608.
[9]. Litjens, G., Kooi, T., Bejnordi, B. E., Setio, A. A. A., Ciompi, F., Ghafoorian, M., ... & Sánchez, C. I. (2017). A survey on deep learning in medical image analysis. Medical image analysis, 42, 60-88.
[10]. Varma, M., Lu, M., Gardner, R., Dunnmon, J., Khandwala, N., Rajpurkar, P., ... & Patel, B. N. (2019). Automated abnormality detection in lower extremity radiographs using deep learning. Nature Machine Intelligence, 1(12), 578-583.
[11]. Wang, S. H., & Zhang, Y. D. (2020). DenseNet-201-based deep neural network with composite learning factor and precomputation for multiple sclerosis classification. ACM Transactions on Multimedia Computing, Communications, and Applications (TOMM), 16(2s), 1-19.
[12]. Naskinova, I. (2023). Transfer learning with NASNet-Mobile for Pneumonia X-ray classification. Asian-European Journal of Mathematics, 16(01), 2250240.
[13]. Albashish, D., Al-Sayyed, R., Abdullah, A., Ryalat, M. H., & Almansour, N. A. (2021, July). Deep CNN model based on VGG16 for breast cancer classification. In 2021 International conference on information technology (ICIT) (pp. 805-810). IEEE.
[14]. Behar, N., & Shrivastava, M. (2022). ResNet50-Based Effective Model for Breast Cancer Classification Using Histopathology Images. CMES-Computer Modeling in Engineering & Sciences, 130(2).
[15]. Kora, P., Ooi, C. P., Faust, O., Raghavendra, U., Gudigar, A., Chan, W. Y., ... & Acharya, U. R. (2022). Transfer learning techniques for medical image analysis: A review. Biocybernetics and Biomedical Engineering, 42(1), 79-107.
[16]. Xu, H., Park, S., Lee, S. H., & Hwang, T. H. (2019). Using transfer learning on whole slide images to predict tumor mutational burden in bladder cancer patients. BioRxiv, 554527.
[17]. Nobashi, T., Zacharias, C., Ellis, J. K., Ferri, V., Koran, M. E., Franc, B. L., ... & Davidzon, G. A. (2020). Performance comparison of individual and ensemble CNN models for the classification of brain 18F-FDG-PET scans. Journal of Digital Imaging, 33, 447-455.
[18]. Le, M. H., Chen, J., Wang, L., Wang, Z., Liu, W., Cheng, K. T. T., & Yang, X. (2017). Automated diagnosis of prostate cancer in multi-parametric MRI based on multimodal convolutional neural networks. Physics in Medicine & Biology, 62(16), 6497.









