1. Introduction
The advent of precision medicine has transformed healthcare delivery, with personalized drug delivery systems (PDDS) assuming a critical role in modern medical practice. Conventional drug delivery systems often adopt a "one-size-fits-all" approach, disregarding individual patient variability, which may lead to suboptimal therapeutic outcomes or unnecessary side effects. The rapid advancement of machine learning techniques has introduced novel strategies to address these challenges.
This study aims to develop a machine learning-based prediction model for personalized drug delivery. By analyzing multidimensional medical data-including patient demographics, clinical indicators, and treatment records, combined with modern machine learning algorithms, we seek to customize optimal drug delivery strategies for individual patients. This approach not only enhances therapeutic efficacy but also reduces the risk of adverse reactions, thereby enhancing the overall quality of healthcare.
The primary objectives of this study are to: (1) establish a reliable patient data analysis framework, (2) develop and optimize machine learning models for predicting personalized drug release curves, and (3) evaluate the performance of different machine learning algorithms in this application.
2. Literature review
The integration of artificial intelligence (AI) and machine learning (ML) into drug delivery systems marks a significant advancement in pharmaceutical technology. The literature review highlights key developments and perspectives in this field.
Kamaly et al. provided a comprehensive review of degradable controlled-release polymers and polymeric nanoparticles, focusing on the mechanisms of drug release control. They emphasized the importance of understanding polymer-drug interactions and release kinetics, which are essential for developing intelligent drug delivery systems and personalized medicine [1]. Schneider et al. introduced AI in drug design, demonstrating its potential to revolutionize traditional drug development by predicting drug-target interactions and optimizing delivery parameters, reducing time and costs while improving accuracy [2].
Hassanzadeh et al. explored how AI can optimize drug delivery system design by adjusting parameters like particle size, drug loading efficiency, and release kinetics. They emphasized the role of machine learning in predicting drug release patterns and personalizing treatments based on patient-specific factors [3]. Gholap et al. reviewed how ML techniques, including deep learning and ensemble methods, can improve drug delivery systems by predicting drug-polymer compatibility and optimizing formulation parameters [4].
Serrano et al. discussed AI's impact on personalizing medicine through improved drug discovery and delivery. They showed how ML can optimize drug delivery parameters based on patient data, leading to more effective treatments. Their work highlighted AI's potential in developing predictive models for individual drug responses [5]. Vora et al. focused on AI’s practical applications in predicting drug release profiles and optimizing delivery system parameters, particularly in creating smart drug delivery systems that can adapt to patient-specific needs [6].
These studies collectively show the progress and potential of AI in drug delivery systems. The literature demonstrates that ML can enhance efficiency and effectiveness while moving towards personalized treatments. However, further research is required to fully realize AI’s potential, especially in validating predictive models for clinical applications. This body of work lays the foundation for our current study, which aims to develop machine learning models to predict personalized drug delivery profiles, contributing to further advancements in the field.
3. Data introduction
This study uses patient data from open medical data sets for analysis. The data set contains 10,000 patient records, covering several key medical characteristic variables. Specifically, the data set contains important indicators such as basic demographic characteristics of patients (such as age and gender), medical insurance type, diagnosis results, hospitalization information (including hospitalization type and length of stay) and medical expenses. All patient data are anonymized to protect patient privacy. The disease diagnosis in the data set covers many common diseases, including cardiovascular diseases, respiratory diseases, digestive system diseases, etc., and has strong representativeness and universality.
In order to ensure the data quality, this paper preprocesses the original data set, including missing value processing, abnormal value detection and processing. In the process of data cleaning, this paper pays special attention to the distribution characteristics of numerical variables (such as medical expenses and hospital stay), and codes and standardizes classified variables (such as insurance types and diagnosis results). After data preprocessing, a complete and reliable analysis sample is finally obtained, which provides a solid data foundation for the training and verification of the subsequent machine learning model.
The dataset's key advantage lies in its rich clinical and treatment-related information, making it highly valuable for developing personalized drug delivery models. By analyzing this multidimensional patient data, we can explore the relationship between individual characteristics and treatment outcomes, thereby supporting the optimization of drug delivery systems.
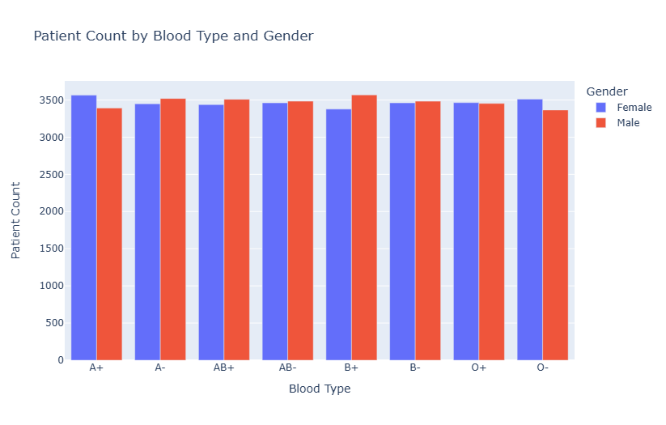
Figure 1: Distribution of patient counts by blood type and gender
Figure 1 displays the distribution of male and female patients across different blood types. The chart reveals fluctuations in patient numbers by gender in each blood group, suggesting a potential correlation between blood type and gender. Notably, blood types like A+ and B+ show a significant disparity between male and female patient counts, indicating that gender may influence blood group distribution, possibly due to genetic or physiological factors.
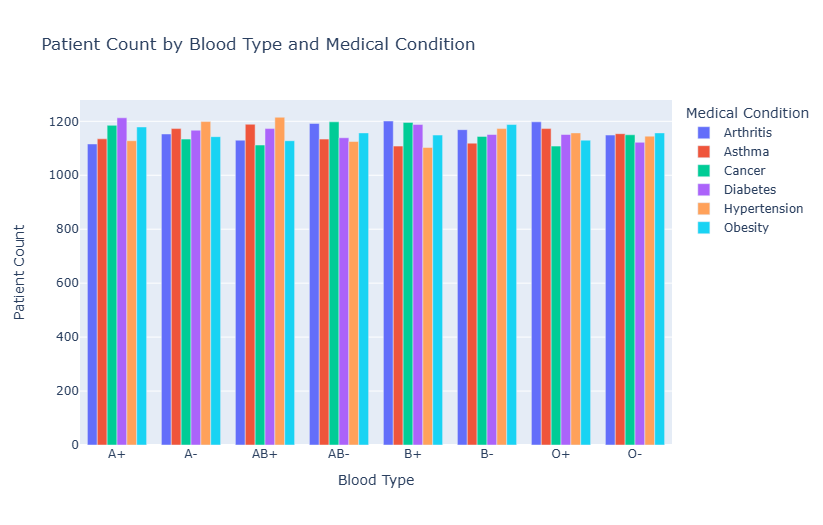
Figure 2: Patient count by blood type and medical condition
Figure 2 shows the distribution of patients with various diseases across different blood types, including arthritis, asthma, cancer, diabetes, hypertension, obesity, and others. The figure reveals uneven disease distribution among blood types. For example, in the A+ blood group, diabetes and hypertension are more prevalent, while in the B+ blood group, asthma and obesity are more prominent. This suggests a potential relationship between blood type and disease susceptibility, indicating that individuals with certain blood types may have a higher predisposition to specific diseases.
4. Model introduction
4.1. XGBoost (eXtreme gradient boosting)
XGBoost is an efficient, distributed gradient boosting decision tree algorithm that improves upon traditional GBDT in both algorithmic design and engineering implementation [7,8]. Its core objective function comprises two components: a training loss term and a regularization term.
\( obj=\sum _{i=1}^{n}l({y_{i}},\hat{{y_{i}}})+\sum _{k=1}^{K}Ω({f_{k}}) \)
Where \( l({y_{i}},\hat{{y_{i}}}) \) is the training loss function, which measures the difference between the predicted value and the real value; \( Ω({f_{k}}) \) is a regularization term used to control the complexity of the model. The regularization term is defined as:
\( Ω(f)=γT+\frac{1}{2}λ|w{|^{2}} \)
Here, \( γT \) controls the number of leaf nodes, and \( λ \) controls L2 regularization of leaf weights. In the t-round iteration, the objective function can be approximated by the second-order Taylor expansion as:
\( {L^{(t)}}=\sum _{i=1}^{n}[{g_{i}}{w_{q}}({x_{i}})+\frac{1}{2}{h_{i}}w_{q}^{2}({x_{i}})]+γT+\frac{1}{2}λ\sum _{j=1}^{T}w_{j}^{2} \)
Where \( {g_{i}} \) and \( {h_{i}} \) are the first and second derivatives of the loss function respectively.
4.2. LightGBM (light gradient boosting machine)
LightGBM is an efficient gradient lifting framework developed by Microsoft. Its main innovation lies in the decision tree algorithm based on histogram and the leaf growth strategy with depth restriction [9]. When the node is split, the gain calculation formula is:
\( Gain=\frac{1}{2}[\frac{G_{L}^{2}}{{H_{L}}+λ}+\frac{G_{R}^{2}}{{H_{R}}+λ}-\frac{{({G_{L}}+{G_{R}})^{2}}}{{H_{L}}+{H_{R}}+λ}]-γ \)
Where \( {G_{L}} \) and \( {G_{R}} \) are the first-order statistics of the left and right child nodes, and \( {H_{L}} \) and \( {H_{R}} \) are the second-order statistics. The formula for calculating the optimal leaf value is:
\( {w^{*}}=-\frac{G}{H+λ} \)
LightGBM greatly reduces the memory consumption and calculation amount through histogram algorithm, and adopts unique feature parallelism and data parallelism methods to improve the training speed.
4.3. CatBoost (categorical boosting)
CatBoost is a machine learning algorithm developed by Yandex, which especially optimizes the processing of category features and introduces sorting promotion to prevent prediction deviation. Its prediction function is:
\( \hat{{y_{i}}}=\sum _{j=1}^{n}{a_{j}}\cdot {Tree_{j}}({x_{i}}) \)
For category features, CatBoost uses an innovative coding method of target statistics:
\( CategoryAvg=\frac{\sum _{i=1}^{n}[{x_{i}}=category]\cdot {y_{i}}+a\cdot P}{\sum _{i=1}^{n}[{x_{i}}=category]+a} \)
Where a is the smoothing parameter and p is the prior probability. The overall loss function of the model is:
\( L(y,f)=\sum _{i=1}^{n}l({y_{i}},f({x_{i}}))+\sum _{j=1}^{n}λ|{f_{j}}{|^{2}} \)
These three ensemble learning algorithms each exhibit unique characteristics and offer distinct advantages in practical applications.
The primary strengths of the XGBoost algorithm lie in three key areas. First, it improves model convergence by approximating the objective function through second-order Taylor expansion. Second, it incorporates a regularization term to control model complexity and reduce the risk of overfitting. Third, XGBoost supports both feature-parallel and data-parallel computation, significantly enhancing computational efficiency.
As a lightweight gradient boosting framework, LightGBM is particularly advantageous in terms of computational performance. It introduces a histogram-based decision tree algorithm that reduces memory usage. Additionally, the leaf-wise growth strategy minimizes computation, and its efficient parallel processing further accelerates training speed.
CatBoost, on the other hand, demonstrates unique advantages in handling categorical features. It introduces an innovative approach for processing categorical variables, improving model performance on such data. Furthermore, it reduces prediction bias through the implementation of ordered boosting. CatBoost also automatically handles missing values and categorical features, thus simplifying data preprocessing and reducing the manual workload.
5. Model results analysis
Figure 4 presents the confusion matrix of the three models, which can directly reflect the prediction accuracy of the models in various categories and show the distribution of real categories and prediction categories.
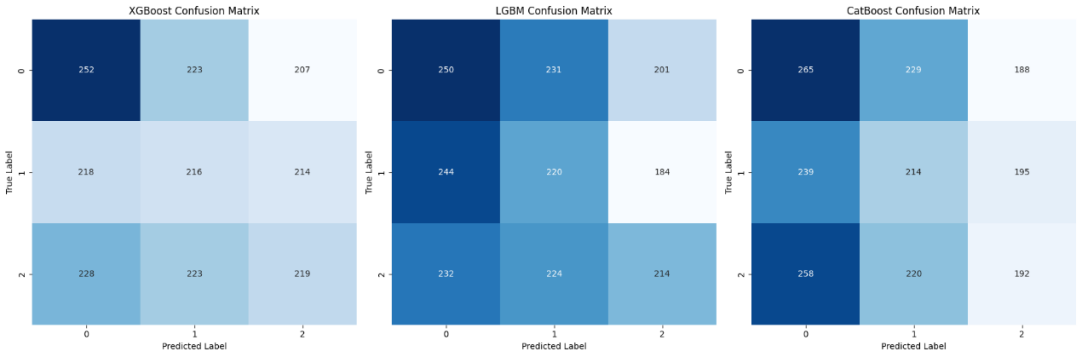
Figure 3: Confusion matrix of classification results
Table 1: Comparison of classification results of different models
Model | Accuracy | Precision | Recall | F1 |
XGBoost Classifier | 0.3435 | 0.3512 | 0.3624 | 0.3473 |
LGBM Classifier | 0.3420 | 0.3489 | 0.3687 | 0.3454 |
CatBoost Classifier | 0.3355 | 0.3401 | 0.3435 | 0.3378 |
According to the data in Table 1, the XGBoost classifier achieved an accuracy of 0.3435, precision of 0.3512, recall of 0.3624, and an F1 score of 0.3473. The LGBM classifier yielded an accuracy of 0.3420, precision of 0.3489, recall of 0.3687, and an F1 score of 0.3454. The CatBoost classifier reported an accuracy of 0.3355, precision of 0.3401, recall of 0.3435, and an F1 score of 0.3378.
Overall, the performance metrics of the three models are relatively close. XGBoost demonstrated slightly higher accuracy and F1 score than the other two models, indicating better overall performance. LGBM achieved the highest recall, suggesting a stronger ability to capture positive instances. In contrast, CatBoost performed slightly weaker across all metrics.
These indicators collectively reflect the models’ classification performance. Accuracy measures the proportion of all correct predictions, precision represents the proportion of correctly predicted positive samples, recall reflects the proportion of actual positives correctly identified, and the F1 score provides a balanced evaluation of both precision and recall.
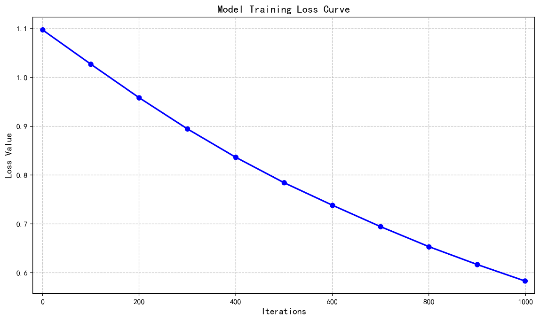
Figure 4: Model training loss curve
Figure 5 shows the training loss curve, illustrating the loss value variation with the number of iterations. Initially, the loss value is around 1.1, indicating poor model fitting. As iterations progress, the loss decreases significantly in the first 200 iterations, showing rapid learning and improvement. Afterward, the decline rate slows, and the loss reaches approximately 0.58 by 1000 iterations. The overall curve is monotonically decreasing, indicating effective error reduction and convergence. However, attention is needed to monitor the performance on the validation set to avoid potential overfitting.
6. Conclusions
This research contributes to the field of personalized drug delivery by developing and implementing machine learning-based predictive models.A comprehensive analysis of patient data, combined with advanced machine learning algorithms, demonstrates substantial potential for improving drug delivery optimization.The comparison of XGBoost, LightGBM, and CatBoost revealed varied strengths in prediction accuracy and recall, with XGBoost exhibiting marginally superior overall performance in accuracy and F1 score.
The training process showed robust convergence, with the loss function consistently decreasing across iterations and stabilizing at an acceptable level.
However, the current accuracy rates suggest room for improvement, potentially through the incorporation of larger datasets and more sophisticated algorithmic approaches. The findings underscore the viability of machine learning applications in personalized medicine while highlighting areas for future enhancement.
Looking ahead, this research opens promising avenues for future exploration, such as expanding data sources, applying deep learning architectures, and conducting clinical validation.
The implications of this work extend beyond theoretical frameworks, offering practical insights for the advancement of precision medicine and personalized therapeutic approaches. Although challenges remain in achieving optimal prediction accuracy, this study provides a solid foundation for ongoing development in personalized drug delivery.
References
[1]. Kamaly N, Yameen B, Wu J, et al. Degradable controlled-release polymers and polymeric nanoparticles: mechanisms of controlling drug release[J]. Chemical reviews, 2016, 116(4): 2602-2663.
[2]. Schneider P, Walters W P, Plowright A T, et al. Rethinking drug design in the artificial intelligence era[J]. Nature reviews drug discovery, 2020, 19(5): 353-364.
[3]. Hassanzadeh P, Atyabi F, Dinarvand R. The significance of artificial intelligence in drug delivery system design[J]. Advanced drug delivery reviews, 2019, 151: 169-190.
[4]. Gholap A D, Uddin M J, Faiyazuddin M, et al. Advances in artificial intelligence in drug delivery and development: A comprehensive review[J]. Computers in Biology and Medicine, 2024: 108702.
[5]. Serrano D R, Luciano F C, Anaya B J, et al. Artificial intelligence (AI) applications in drug discovery and drug delivery: Revolutionizing personalized medicine[J]. Pharmaceutics, 2024, 16(10): 1328.
[6]. Vora L K, Gholap A D, Jetha K, et al. Artificial intelligence in pharmaceutical technology and drug delivery design[J]. Pharmaceutics, 2023, 15(7): 1916.
[7]. Chen T, Guestrin C. Xgboost: A scalable tree boosting system[C]//Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining. 2016: 785-794.
[8]. Chen T, He T, Benesty M, et al. Xgboost: extreme gradient boosting[J]. R package version 0.4-2, 2015, 1(4): 1-4.
[9]. Ke G, Meng Q, Finley T, et al. Lightgbm: A highly efficient gradient boosting decision tree[J]. Advances in neural information processing systems, 2017, 30.
Cite this article
Wang,Y.;Shao,W.;Lin,J.;Zheng,S. (2025). Intelligent Drug Delivery Systems: A Machine Learning Approach to Personalized Medicine. Applied and Computational Engineering,150,205-211.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 3rd International Conference on Software Engineering and Machine Learning
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Kamaly N, Yameen B, Wu J, et al. Degradable controlled-release polymers and polymeric nanoparticles: mechanisms of controlling drug release[J]. Chemical reviews, 2016, 116(4): 2602-2663.
[2]. Schneider P, Walters W P, Plowright A T, et al. Rethinking drug design in the artificial intelligence era[J]. Nature reviews drug discovery, 2020, 19(5): 353-364.
[3]. Hassanzadeh P, Atyabi F, Dinarvand R. The significance of artificial intelligence in drug delivery system design[J]. Advanced drug delivery reviews, 2019, 151: 169-190.
[4]. Gholap A D, Uddin M J, Faiyazuddin M, et al. Advances in artificial intelligence in drug delivery and development: A comprehensive review[J]. Computers in Biology and Medicine, 2024: 108702.
[5]. Serrano D R, Luciano F C, Anaya B J, et al. Artificial intelligence (AI) applications in drug discovery and drug delivery: Revolutionizing personalized medicine[J]. Pharmaceutics, 2024, 16(10): 1328.
[6]. Vora L K, Gholap A D, Jetha K, et al. Artificial intelligence in pharmaceutical technology and drug delivery design[J]. Pharmaceutics, 2023, 15(7): 1916.
[7]. Chen T, Guestrin C. Xgboost: A scalable tree boosting system[C]//Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining. 2016: 785-794.
[8]. Chen T, He T, Benesty M, et al. Xgboost: extreme gradient boosting[J]. R package version 0.4-2, 2015, 1(4): 1-4.
[9]. Ke G, Meng Q, Finley T, et al. Lightgbm: A highly efficient gradient boosting decision tree[J]. Advances in neural information processing systems, 2017, 30.









