1. Introduction
Cancer is one of the leading causes of death according to the World Health Organization (WHO), with more than two people dying of skin cancer every hour in the United States alone. About 3 million cases of non-melanoma skin cancer and 132,000 cases of melanoma skin cancer are diagnosed worldwide each year. Early diagnosis of skin cancer is an important chance to successfully treat and prevent metastasis to other organs. The 5-year survival rate of skin cancer is 99% when detected early [1]. Therefore, if abnormal growth of skin tissue is suspected, it is very important to segment it as soon as possible, which can directly or indirectly reduce mortality. Skin cancer image segmentation using computer-aided system can better extract features to separate the parts with special meaning in medical images, which is an important technical premise for treatment, and the accuracy of segmentation results directly affects the subsequent treatment.
In practice, the traditional methods of medical image segmentation include threshold method[2], region growth method[2] and clustering method[2]. The most commonly used clustering algorithm in the field of medical segmentation is fuzzy C-mean algorithm (FCM), which combines fuzzy set theory with clustering algorithm to determine the degree of pixel belonging to a certain cluster through "membership degree", which alleviates the problem of few labels in medical image segmentation to a certain extent. Medical image segmentation based on deep learning are Fully Convolutional Networks (FCN) [3] and U-Net network structures[4]. The disadvantage of FCN model is that the global context information is not taken into account and the up-sampling operation is only used once, so details are easily neglected. In view of the shortcomings of FCN, Ronneberger et al. proposed U-Net network structure, which has good performance and is more suitable for medical image segmentation. The automatic segmentation of medical images based on deep learning has been playing an important role in the medical field due to its high speed and high accuracy.
In this paper, we use U-Net network to segment dermatoscopic images of pigmented lesions.
The system is experimented with ISIC dataset to segment malign and benign moles, The experimental results show the precision score, respectively. The analyzation results demonstrate that our skin lesion segmentation system can obtain high accuracy and efficiency in skin cancer segmentation, and provide timely feedback information.
2. Literature review
In clinical diagnosis, it is an important content to diagnose skin lesions according to color dermatoscopic images. However, due to the limited availability of dermatologists, early detection of melanoma is often delayed. At the same time, due to the influence of skin surface hair, color, blood vessels and low contrast between diseased skin and diseased skin, even experienced clinicians cannot accurately determine the lesion area on the skin, so as to fail to correctly diagnose malignant melanoma. At this time, the Computer Aided Diagnostic system (CAD) [5] can effectively segment the skin lesion area, thus improving the diagnosis rate of malignant melanoma. In order to ensure the normal operation of CAD in detecting melanoma in cutaneous microscopic images, it is very important to accurately detect the boundary of lesions. The reasons are as follows: in order to properly analyze the indicators existing in the lesions, it is necessary to carry out appropriate segmentation; Secondly, some values calculated according to the boundary itself, such as the degree of division in "ABCD method" and "Menzies method" [6], are themselves diagnostic indicators of malignant tumors. Skin lesion Image segmentation can be divided into manual segmentation and automatic segmentation. Manual segmentation is performed by clinicians based on their own pathological knowledge of skin lesions with the help of Image instance segmentation annotation tools. The lesion area on the skin lesion image was manually divided. Automatic image segmentation methods mainly include two kinds of methods: one is the traditional image segmentation algorithm, the other is based on deep learning segmentation algorithm. In the field of medical image segmentation, Ronneberger proposed a new convolutional neural network segmentation method, U-Net When training the network, U-Net can use ResNet, VggNet and other pre-training network weight parameters, which can greatly reduce the training time. Another feature of U-Net is that the feature maps obtained from each convolution layer are concatenate to the corresponding up sampling layer through skip-connection, so that the feature maps of each layer can be effectively used in subsequent calculations. U-Net has become a medical image by virtue of its excellent performance in segmentation.
Based on the U-Net framework, many semantic segmentation networks with superior performance in different medical images are derived.
U-net++ [7]is one of the most important improvements to U-Net. The main change is to change U-Net's jump connection structure to dense connection. Through short-link and up-down sampling operations, the features of multiple different levels are indirectly fused. Therefore, decoder can perceive objects of different sizes under different sensing fields, which plays a good role in the segmentation effect of different parts of medical images.
Res-U-Net[8] and Dense-U-Net[9] are inspired respectively by residual connection and dense connection, and each sub module of U-Net is replaced by a form with residual connection and high- density connection respectively. Res-U-NET is used in retinal image segmentation. A high-density connection is part of a combination of inputs from the next layer and outputs from the previous layer. All submodules of U-Net will be replaced by high-density U-Net modules, and it is recommended to use fully high-density U-Net modules to eliminate the falsity on the images.
Attention U-Net [10] adopts an attention mechanism in U-Net to adjust the output fit degree of the encoder and combine the resolution fit degree of the encoder with the absolute fit degree of the decoder. It is noteworthy that in the long-term domain, the target has the tendency to obtain large vocabulary, and in the background domain, the target has the tendency to obtain small difference vocabulary, especially the high accuracy of image segmentation.
Some medical images are stored in the form of 3D, such as CT and MRI images. 3D-U-Net network is used to segment such images, replacing 2D convolution operation with 3D convolution operation, and introducing a new objective function into the model to effectively alleviate the problem of class imbalance in image segmentation. Wang Jiping et al. [11] used 3D-U-Net to perform automatic segmentation of organs at risk for nasopharyngeal carcinoma, which saved 74.5% of the average time compared with manual drawing, and the accuracy was higher than 80%.
U-Net is a classical network design method, which is widely used in image segmentation. There are also many new ways to improve on this foundation, incorporating newer web design concepts.
3. Method
3.1. U-Net Structure
The network is basically a symmetrical structure, with encoder on the left and decoder on the right. The network is composed of 3x3 convolution layer, Relu function, maximum pooling layer window size of 2x2, step size of 2, up sampling convolution and Concatenation.
The feature number of each down sampling is doubled, and the feature number of each up sampling is halved.
The most important feature of the network is that the splicing operation is added, because the image segmentation needs both global location information and context information. Feature abstraction is carried out by down sampling to obtain global location information, but the context information of small feature maps will be lost. Therefore, in the process of up-sampling, the method of splicing is used.
The best parts of U-Net are these three parts: up sampling, down sampling and skip connection.
U-Net performs a total of 4 up-sampling sessions in the same phase using Skip Connection. In this way, the final recovered feature pattern will become the feature of the lower stage, instead of directly monitoring the higher semantic feature, and the loss will be reversed. At the same time, the features of different scales are integrated to realize multi-scale prediction and depth monitoring. After four times of sampling, the edge information of the subdivided image is restored. The parts before pool4 are all down sampling. The size of the image input is (512, 512, 1).
The parts after the pool5 layer are all up sampling, and the shallow features extracted by pool1-pool4 are combined in the up sampling process. The concatenate function used here is used to combine the matrices.
Finally, a 1*1 scale convolutional layer is used, combined with the softmax function for classification.
3.2. Image Processing
Running data.py generates three npy files: Imgs_mask_train. npy is the training picture tag, Imgs_train. npy is the training picture and Imgs_test. npy is the test image. This test picture is the final picture to be tested, rather than the picture of test accuracy in the training. The picture of test accuracy and the training picture are together. During the training, part of the training picture will be allocated to test accuracy with a probability of 4 to 1.
3.3. Experiment Settings
The experiments in this chapter are based on TensorFlow deep learning framework in Windows system, and the processor is IntelCoreTMi7 6700CPU@340GHz*8. The algorithm is written in Python language. The network is trained by two NVIDIA GeForcegTX1070 Gpus with 8G memory each, and CUDA version is 90. The network weight parameters were initialized by Xavier, and the optimizer was Adam, which accelerated the convergence speed of the objective function. The epoch size of network training was set to 10, batchsize was set to 2 during network training, and the image resolution was converted to 512x512. During the test, Batchsize is set to 1, and the test images are segmented one by one. Finally, the segmented images are saved and relevant evaluation indicators are calculated.
4. Results
4.1. Dataset
In my experiment of Skin Image Segmentation Module, I use images from the dataset: Skin Imaging Collaboration: Melanoma Project (ISIC).
ISIC is an academic and industry partnership to promote digital skin imaging applications to reduce melanoma mortality. It consists of a database containing 13,786 skin endoscopy images (as of February 12, 2018) classified as malignant and benign skin lesions [4]. Each example contains an image of the lesion, metadata about the lesion including classification and segmentation. As a result of that, I use this dataset to train my U-Net model which is built to segment malign and benign melanocytic lesions.
4.2. Skin Image Analysis Module Performance
Running data.py generates t
In the experiment of Skin Image Analysis Model, images were randomly selected to train the model, and the accuracy of the model was tested in the remaining images. The ratio of the number of images in the two parts was 4:1. For example, if 30 images are selected from the data set as the training set, 24 images are randomly selected to train the model, and the remaining 6 images are tested for accuracy. If 100 images are selected from the data set as the training set, 80 images are randomly selected to train the model, and the remaining 20 images are tested for accuracy.
Due to the different sizes of the original data images in ISIC, the data is preprocessed and the unified pixel is 512*512, which is convenient to input into the network for training.
During the training process, the number of training iterations is set to epoch=10, that is, all samples in the training set are trained ten times in total. Set batch size=2, that is, 2 samples are taken in the training set for each training, and 40 training sessions are required for each iteration of 80 images .
In the experiment of setting 1, the network is trained by CPU, and the training set contains 30 images. In the experiment of setting 2 and setting 3, the network is trained by two NVIDIA GeForcegTX1070 GPUs. Training set in setting 2 still contains 30 images, while setting 3 has 100 images.
After 10 epochs of training iteration, the training accuracy of setting 3 is about 0.9085, while the validation accuracy is about 0.9536, as shown in figure 5. As a result, the model gives accurate feedback in learning and analysis in time.
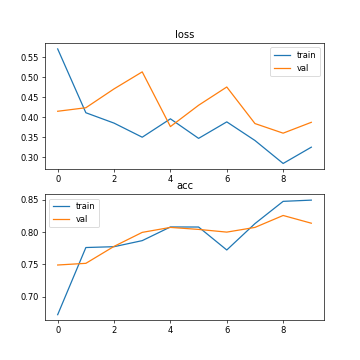
Fig. 1. The accuracy and loss (setting 1: 30 training images, CPU running, epoch=10, batch size=2).
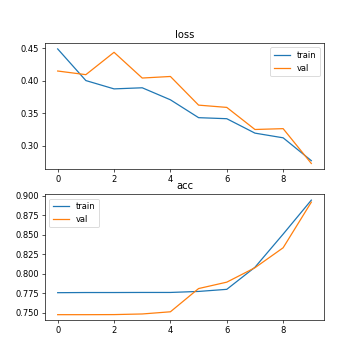
Fig. 2. The accuracy and loss (setting 2: 30 training images, GPU running, epoch=10, batch size=2).
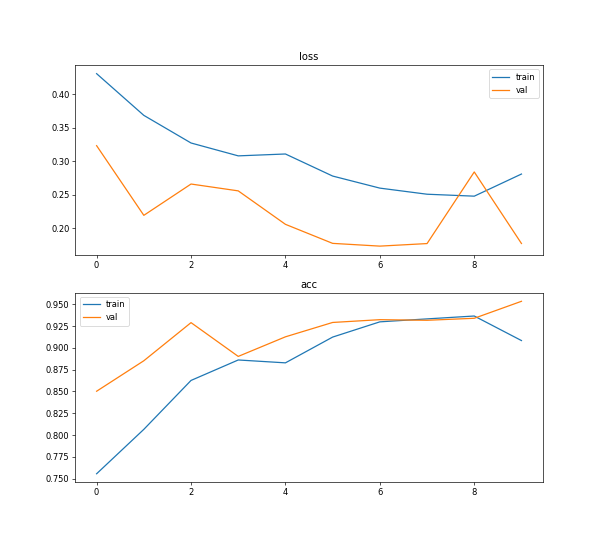
Fig. 3. The accuracy and loss (setting 3: 100 training images, GPU running, epoch=10, batch size=2).
4.3. Skin Image Segmentation Module Performance
Under different numbers of training dataset, the accuracy rate and loss of the model are different. There is also a difference between GPU training and CPU training models. See the following table.
segmentation performances
Accuracy | Validation accuracy | Loss | Validation loss | Running time of each epoch | |
Setting1 (CPU running, 30 training images, epoch=10, batch size=2) | 0.8495 | 0.8137 | 0.3253 | 0.3874 | 312s |
Setting2 (GPU running, 30 training images, epoch=10, batch size=2) | 0.8947 | 0.8921 | 0.2770 | 0.2727 | 292s |
Setting3 (GPU running, 100 training images, epoch=10, batch size=2) | 0.9085 | 0.9536 | 0.2812 | 0.1776 | 1188s |
TestU-Net.py will test the image that put in the test folder, and the results will be saved in the Results folder. The processing effect after data enhancement is shown in the figure 4 below. After GPU training with 30 images and GPU training with 100 images, the results of model segmentation is shown below. When the training dataset is small, the segmentation result graph is fuzzy.

(a) (b)


(c) (d)
Fig. 4. Segmentation results (setting: 10 epochs, batch size=2). (a) Original image; (b) Segmentation effect of setting 1; (c) Segmentation effect of setting 2; (d) Segmentation effect of setting 3;
5. Discussion
This experiment was originally run using CPU, but PyCharm still showed that the kernel had hung when running the first epoch, and this problem still existed after two or three reboots. Therefore, GPU was used to run the experiment later.
As can be seen from the table, the processing speed of deep learning by CPU is slower than that of GPU. Using GPU training can increase the amount of training data and obtain higher accuracy. In addition, the accuracy of training and testing varies with the number of training samples. The number of training samples increases and the accuracy rate increases. In addition, it can be seen from Figure 5 that after ten iterations, the curve of 10 epoch has tended to be stable, and the sufficient number of training samples saves the number of training times.
In short, CPU is good at complex operations such as global domination, while GPU is good at simple repetitive operations on big data. The CPU is a teaching aid for complex mental work, while the GPU is a manual worker for massive parallel computing [12]. Deep learning is a mathematical network model established to simulate human brain nervous system. The biggest characteristic of this model is that it needs big data to train. Therefore, a large number of parallel and repeated calculations are required for computer processors. Therefore, GPU is more suitable for deep learning.
The number of parameters of the original U-Net is about 28M (the number of parameters of U-Net with up-sampling transpose convolution is about 31M), while the model can be smaller if the number of channels is multiplied. The number of U-Net parameters was 7.75M after being reduced by two times. By four times, the number of model parameters can be reduced to less than 2M, which is very light [13]. Therefore, the number of training samples of U-Net model should not be too large, which is easy to lead to over-fitting.
Due to the fuzzy boundary and complex gradient of medical image, more high-resolution information is needed. At the same time, the internal structure of the human body is relatively fixed, the distribution of segmentation targets in the human body image is very regular, the semantics are simple and clear, and the low-resolution information can be easily located. Therefore, U-Net structure is suitable for medical image segmentation task.
6. Conclusion
In this paper, a skin cancer segmentation system using U-Net is proposed to separate areas of skin tumor from healthy skin, with two main components, Skin Image Analysis Module and Skin Image Segmentation Module. For each module in U-Net, accurate results are displayed in learning and analysis, and segmentation results are returned in time. Under different numbers of training dataset, the accuracy rate and loss of the U-Net model are different. There is also a difference between GPU training and CPU training U-Net models. After 10 epochs of training, the training accuracy of the experiment with CPU running and 30 images running is around 0.8. The training accuracy of the experiment with GPU running and 30 images running is around 0.8. The training accuracy of the experiment with GPU running and 100 images training is around 0.9085, while the validation accuracy is around 0.9536. The processing speed of deep learning by CPU is slower than that of GPU and the accuracy rate increases when the number of training images increases.
In the future work, there are two aspects of improvement. First, the current model uses a fixed size training suite, and developers need to pre-process the image of the training suite to change the resolution. This process can take time to process a large amount of data, so other neural networks can be added to automatically perform the process of changing image resolution. Another improvement is a sharper picture of the results. It is necessary to improve the quality of the image by processing the segmentation result, such as enhancing contrast, sharpening, removing blur and other operations. This will require extra code to magnify the image's grayscale range, making the image sharper.
References
[1]. Abuared,N., Panthakkan,A., Al-Saad,M., Amin,S.A. and Mansoor,W. (2020) Skin Cancer Classification Model Based on VGG 19 and Transfer Learning. In: 2020 3rd International Conference on Signal Processing and Information Security (ICSPIS). pp. 1-4.
[2]. Chulan Ren, Ning Wang, Yang Zhang. Review of medical image segmentation methods [J]. Network Security Technology & Application,2022(02):49-50.
[3]. Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation[C]. Proceedings of the IEEE conference on computer vision and pattern recognition, 2015: 3431-3440.
[4]. Ronneberger O, Fischer P, Brox T.U-net: Convolutional Networks for Biomedical Image Segmentation[C]//International Conference on Medical Image Computing and Computer-Assisted Intervention.Munich: MICCA, 2015: 234-241.
[5]. Ogawa K , Fukushima M , Kubota K , et al. Computer-aided diagnostic system for diffuse liver diseases with ultrasonography by neural networks[J]. IEEE Transactions on Nuclear Science, 2002, 45(6):3069-3074.
[6]. Johr R H . Dermoscopy: alternative melanocytic algorithms—the ABCD rule of dermatoscopy, menzies scoring method, and 7-point checklist[J]. 2002, 20(3):0-247.
[7]. Zhou Z , Siddiquee M M R , Tajbakhsh N , et al. UNet++: A Nested U-Net Architecture for Medical Image Segmentation[C]// 4th Deep Learning in Medical Image Analysis (DLMIA) Workshop. 2018.
[8]. Xiao, Xiao, Shen Lian, Zhiming Luo, and Shaozi Li. “Weighted Res-U-Net for High-Quality Retina Vessel Segmentation.” In 2018 9th International Conference on Information Technology in Medicine and Education (ITME), pp. 327-331. IEEE, 2018.
[9]. Guan, Steven, Amir Khan, Siddhartha Sikdar, and Parag V. Chitnis. “Fully Dense U-Net for 2D Sparse Photoacoustic Tomography Artifact Removal.” arXiv preprint arXiv:1808.10848 (2018).
[10]. Oktay, Ozan, Jo Schlemper, Loic Le Folgoc, Matthew Lee, Mattias Heinrich, Kazunari Misawa, Kensaku Mori et al. “Attention U-Net: learning where to look for the pancreas.” arXiv preprint arXiv:1804.03999 (2018).
[11]. Jiping Wang,Xin Li,Chuanxi Chen, etc. Application of 3D U-Net in automatic segmentation of organs at risk in nasopharyngeal carcinoma [J]. Chinese Medical Equipment Journal,2020,41(11):17- 20+45
[12]. Lianbo Zhong. Comparison of GPU and CPU [J]. Technology and Market, 2009, 16(009):13-14.
[13]. Atlas P . Section of Biomedical Image Analysis (SBIA).
Cite this article
Wang,D. (2023). Skin lesion segmentation of dermoscopy images using U-Net. Applied and Computational Engineering,6,840-847.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 3rd International Conference on Signal Processing and Machine Learning
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Abuared,N., Panthakkan,A., Al-Saad,M., Amin,S.A. and Mansoor,W. (2020) Skin Cancer Classification Model Based on VGG 19 and Transfer Learning. In: 2020 3rd International Conference on Signal Processing and Information Security (ICSPIS). pp. 1-4.
[2]. Chulan Ren, Ning Wang, Yang Zhang. Review of medical image segmentation methods [J]. Network Security Technology & Application,2022(02):49-50.
[3]. Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation[C]. Proceedings of the IEEE conference on computer vision and pattern recognition, 2015: 3431-3440.
[4]. Ronneberger O, Fischer P, Brox T.U-net: Convolutional Networks for Biomedical Image Segmentation[C]//International Conference on Medical Image Computing and Computer-Assisted Intervention.Munich: MICCA, 2015: 234-241.
[5]. Ogawa K , Fukushima M , Kubota K , et al. Computer-aided diagnostic system for diffuse liver diseases with ultrasonography by neural networks[J]. IEEE Transactions on Nuclear Science, 2002, 45(6):3069-3074.
[6]. Johr R H . Dermoscopy: alternative melanocytic algorithms—the ABCD rule of dermatoscopy, menzies scoring method, and 7-point checklist[J]. 2002, 20(3):0-247.
[7]. Zhou Z , Siddiquee M M R , Tajbakhsh N , et al. UNet++: A Nested U-Net Architecture for Medical Image Segmentation[C]// 4th Deep Learning in Medical Image Analysis (DLMIA) Workshop. 2018.
[8]. Xiao, Xiao, Shen Lian, Zhiming Luo, and Shaozi Li. “Weighted Res-U-Net for High-Quality Retina Vessel Segmentation.” In 2018 9th International Conference on Information Technology in Medicine and Education (ITME), pp. 327-331. IEEE, 2018.
[9]. Guan, Steven, Amir Khan, Siddhartha Sikdar, and Parag V. Chitnis. “Fully Dense U-Net for 2D Sparse Photoacoustic Tomography Artifact Removal.” arXiv preprint arXiv:1808.10848 (2018).
[10]. Oktay, Ozan, Jo Schlemper, Loic Le Folgoc, Matthew Lee, Mattias Heinrich, Kazunari Misawa, Kensaku Mori et al. “Attention U-Net: learning where to look for the pancreas.” arXiv preprint arXiv:1804.03999 (2018).
[11]. Jiping Wang,Xin Li,Chuanxi Chen, etc. Application of 3D U-Net in automatic segmentation of organs at risk in nasopharyngeal carcinoma [J]. Chinese Medical Equipment Journal,2020,41(11):17- 20+45
[12]. Lianbo Zhong. Comparison of GPU and CPU [J]. Technology and Market, 2009, 16(009):13-14.
[13]. Atlas P . Section of Biomedical Image Analysis (SBIA).









