1. Introduction
Kidney serves as indispensable component in the elimination of waste substances from blood and the equilibrium of water as well as essential minerals within the human body [1]. The importance of kidney cannot be overstated, as impaired kidney function can result in both significant suffering and mortality; notably, Chronic Kidney Disease (CKD), a type of abnormality of the kidney, has ascended to be the 16th principal cause of mortality, impacting an estimated population of over 800 million individuals worldwide [2]. Predominant kidney abnormalities include the formation of kidney cyst, stone, and tumor [3]. Therefore, kidney cysts are fluid-filled sacs, which form on the surface of the kidney [4]. Kidney stones are crystalline concretion of minerals that develop within the kidney. They are closely associated with end-stage renal failure, impacting approximately 12% of the global population [5]. In terms of kidney tumor, it can be further categorized into two different phenotypes: benign tumor and malignant kidney tumor, also known as renal cell carcinoma (RCC). RCC has emerged to be one of the ten most common cancers on a global scale [4, 6].
The early stages of kidney abnormality often manifest without symptoms and gradually progress over the course of several years until reaching the end stage [7]. The principal treatment of end-stage renal failure is renal replacement, which typically incurs a significant financial burden for the majority of patients [8]. Therefore, early-stage detections and interventions are crucial in found in preventing and delaying the progression of kidney abnormalities [8]. Computed tomography (CT) imaging is highly preferred by radiologists in diagnosing kidney abnormality due to its capability to produce high-resolution images characterized by clear anatomical details, optimal contrast, and superior spatial resolution [4]. However, radiologists and nephrologists may occasionally encounter challenges in accurately diagnosing certain cases, leading to the possibility of misdiagnosis [9]. Furthermore, the current standard CT screening procedures are frequently associated with exorbitant financial costs [10].
In recent times, deep learning emerged as a highly efficient and cost-effective approach for ameliorating kidney disease diagnosis via image processing and classification. Collecting medical images, nevertheless, poses significant challenges for scientists [11]. Ethical issues such as safeguarding patients’ privacy, ensuring anonymity, and obtaining informed consent are closely associated with the data-gathering process [11]. Various laws and regulations are set to ensure the ethical standards are compiled during all process of data collection. As a result, the availability of medical data is relatively limited compared to other fields. Furthermore, the situation is exacerbated by the lack of sufficient public medical image datasets, despite the existence of several published researches in related fields [4].
Transfer learning offers an effective solution by leveraging the ability to transfer learned parameters from pre-trained Convolutional Neural Network (CNN) models trained on large datasets [12]. Specifically, transfer learning involves two approaches: feature extraction and fine-tuning [12]. ImageNet is a large database that consist of more than 14 millions of annotated images often used in transfer learning [12]. The utilization of ImageNet in transfer learning, which encompasses diverse images across various domains, enables a pre-trained model to learn shapes similar to those in the target domain through images belonging to other fields [12]. However, it is worth nothing that the VGG-16 model’s pre-trained weights are derived from training on the ImageNet dataset, which primarily consists of general images rather than medical images. This issue also arises with transfer learning-based algorithms for other medical data set recognition [13-15]. Therefore, the extent to which these pre-trained weights can be effectively transferred and applied to medical imaging tasks requires further investigation and exploration. Very Deep Convolutional Networks (VGG) is a type of CNN model that is one of the best computer vision models [16]. VGG can be trained on ImageNet, which allows it to transfer pre-trained weights from general images to specific target models. Despite its simplicity, VGG has been proven to be able to classify 1000 images in 1000 distinctive domains with an accuracy of 92.7% [16]. Such performance highlights the effectiveness and reliability of VGG for image classification tasks, further supporting its suitability for application in the classification of Kidney CT Images.
In this study, two VGG16 models were constructed and compared. One model had randomly initialized weights, while the other model utilized ImageNet as the pre-training dataset. The evaluation was performed on a publicly available online dataset [17]. The performance of each model was evaluated and compared using metrics such as accuracy score and loss rate. However, solely relying on these numbers does not offer a comprehensive understanding of the underlying reasons for a model’s success or underperformance. To gain a deeper insight into the models’ performance, this study employed techniques such as Grad-CAM, saliency maps, and confusion matrix to provide explanations and visualization that shed light on the factors contributing to the models’ performance.
The remaining sections of this paper are organized as follows: Section 2 presents a description of the dataset and the approach. Section 3 presents the results and provides an analysis. Section 4 discusses the conclusion, along with potential future works.
2. Methods
2.1. Dataset preparation
This study uses a publicly available dataset found on Kaggle [17]. This dataset contains 12,446 distinctive RGB-based CT image data in different size divided into four unique categories: cysts, normal (cases), (kidney) stones, and tumors. Specifically, the dataset contains 3,709 cysts, 5,077 normal cases, 1,377 kidney stones, and 2,283 tumors [18]. These data were gathered with the help of Mehedi Hasan, a Medical Technologist, from multiple hospitals in Bangladesh. Figure 1 provides a sample CT image with tumor.
Figure 1. A sample CT image with Tumor.
2.2. CNN-based kidney CT images classification
As a type of deep learning model that can extract key features through convolving grid-like topology, CNN is especially suitable for processing images [18]. What sets CNN apart from other deep learning models is its capability to learn and extract the unique features and patterns within images. In a conventional CNN architecture, the initial component is the convolutional layer, succeeded by a pooling layer, and ultimately a fully connected layer [19] (shown in Figure 2). Typically, a convolutional layer is paired with a pooling layer, performing the feature extraction process of a CNN [19]. The output from the previous layers is subsequently mapped to the final output layer through a fully connected layer.
The convolution layer processes the images with a kernel, which is an array of numbers. At every position of the input, the kernel's individual elements are multiplied with the corresponding elements of the input. The resulting products are then summed together to calculate the value of that specific position in the output. Since features can be located anywhere within the input image, the kernel needs to traverse the entire image. This movement of the kernel across the image is referred to as convolution, and the step size by which the kernel moves is known as the stride. After the entire input image has been convolved, an output is generated consisting of all the summed products between the input and the kernel. This output is referred to as the feature map [18].
To minimize the number of learned parameters, which is also known as the dimensionality, pooling layer provides two different pooling methods: global average pooling and max pooling. As suggested by the name, global average pooling is to take the average value of all components in the feature maps whereas the maximum value of each patch is chosen in max pooling. The most significance difference between the two pooling methods is that global average pooling can reduce the feature map’s size down to 1x1, but in the other case the size remains the same [18].
Ultimately, the pooled feature maps are flattened to facilitate output computation. Connecting pooled features to neurons in the fully connected layer, the final result is produced [18].
VGG Very Deep Convolutional network (VGG) is published by Simonyan and et al. in 2015 [19]. Compared to other CNN architecture, VGG is known for its higher level of depth, typically 16 to 19 layers. The depth allows for a more complex and expressive feature representation. Additionally, VGG is characterized by its very small (3x3) convolution filters followed by 2x2 max-pooling layers. This is useful for constructing deeper architecture and simplifies the implementation process [19, 20]. A notable advantage of VGG is its ability to leverage pre-trained weights on large scale dataset, providing transferability to VGG. By specifying ImageNet as the dataset for the pre-trained weight to be trained on, the model can benefit from the knowledge and feature representation learned from vast amounts of data. Alternatively, if no pre-trained weights are specified, a random large-scale dataset would be used to initialize the weights [20].
In this study, the CNN models are constructed based on VGG-16 architecture due to lack of sufficient amount of medical image data. There are 1000 neurons in the output layer in the VGG-16 model originally, corresponding to the 1000 categories in the dataset of that publication; however, in this study, there are only four classes, so the output layer is reduced to four neurons in order to match the number of categories. This study focuses on the difference between performances of VGG-16 models trained with ImageNet and without ImageNet; hence, two identical models are constructed. The only difference is that one model has weights specified to ImageNet and the other one is set to none.
The evaluation of model performance typically involves assessing test accuracy, which provides an indication of the model's classification capabilities but lacks explanatory insights. Therefore, saliency maps and Grad-CAM are used in this study to provide a more comprehensive insight into the performance of the models. Saliency map highlights the features and areas that impact the prediction the most with bright pixels. On the other hand, Grad-CAM is a technique to generate class specific heat map, which can also show the features that are extracted from the feature map.
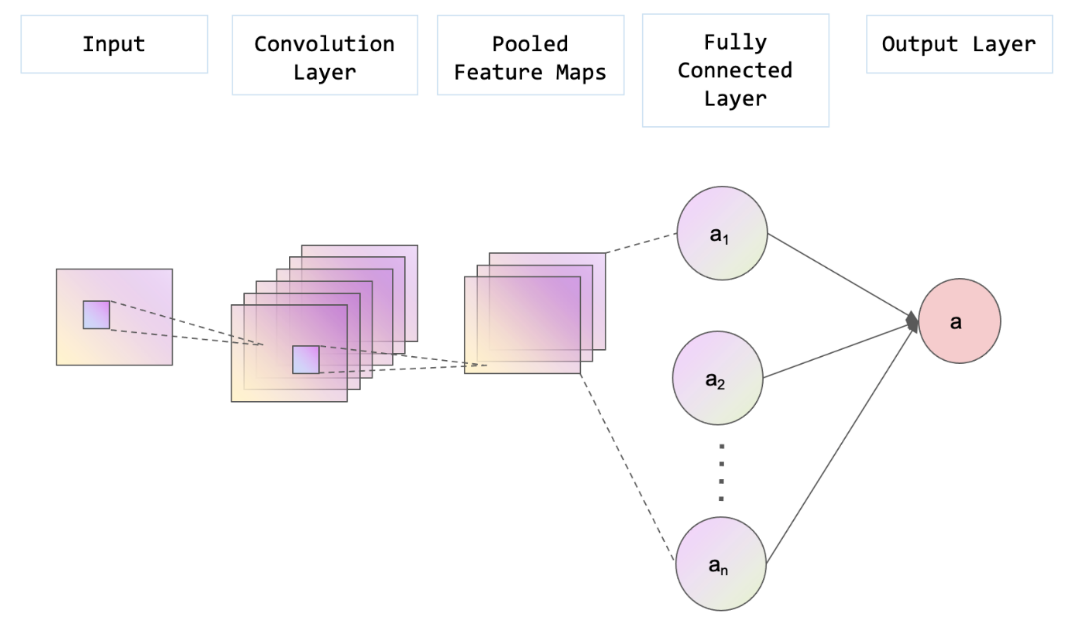
Figure 2. The structure of a CNN (Photo/Picture credit: Original).
2.3. Implementation details
In this study, the TensorFlow framework was utilized for training the model. To analyze the performance of the VGG-16 models trained on ImageNet and other datasets, two CNN models were constructed: one with the weights set to ImageNet, and the other with weights set to none. Both models share the same structure, which begins with a VGG-16 model. The input size for the model is set to 200x200 pixels. The flatten layer is concatenated to the VGG-16 model to reshape the image for further processing. Subsequently, a dense layer with 1024 neurons is incorporated, employing the Rectified Linear Unit (ReLU) activation function. Finally, the output layer consists of four neurons, corresponding to the four distinct labels, with the activation function set to SoftMax as the model performs multi-class classification.
In consideration of the multi-class classification nature of the models, the chosen loss function for this study is categorical cross-entropy. The models were compiled using the Adam optimizer and accuracy was selected as the evaluation metric. To mitigate the overfitting problem on the training set, an early stopping callback was employed. To reduce training time while still capturing important patterns, 30 epochs were run during the training process. The batch sizes were 100 which is identical for training, validation, and test sets.
3. Results and discussion
There is a significant performance gap between the models trained with ImageNet and the one trained with a random dataset. The accuracy of the test set for the ImageNet-trained model reaches 98.96%, whereas the test accuracy of the model trained with the random dataset merely achieves 68.94%. These discrepancies are further evident when comparing the confusion matrices, saliency map, and Grad-CAM. Ideally, all predictions should lie into only the grids along the diagonal line of the confusion matrix, representing true positive predictions, indicating 100% accurate prediction. However, in the confusion matrix of the model trained without ImageNet (Figure 3), a large number of predictions lie into grids that are not on the diagonal line, which represents a significant number of misclassifications. In contrast, the confusion matrix of the ImageNet-trained model (Figure 4) exhibits a higher accuracy, with most of the prediction lines aligning closely along the diagonal.
To gain deeper insights into these performance differences, saliency maps and Grad-CAM can be used to identify the features that contribute most to the predictions. In the saliency map of the model trained without ImageNet (Figure 5), light points are dispersed throughout the entire image, making it challenging to pinpoint any specific feature of significance. This suggests that the model might not have adequately learned the relevant features during training, resulting in a lower overall accuracy rate. Similarly, the Grad-CAM for this model (Figure 5) does not highlight any specific areas, indicating a lack of feature recognition by the model. On the other hand, both the saliency map (Figure 6) and Grad-CAM (Figure 6) of the ImageNet-trained model exhibit highlighted areas that the model focuses on during predictions. These findings suggest that the model has successfully learned and can rely on certain discernible features in the images, contributing to its higher accuracy.
Based on the evidence provided above, it can be concluded that incorporating ImageNet as a pre-training dataset has notably enhanced the model's performance and its ability to make accurate predictions. One plausible explanation for this improvement is that even though ImageNet comprises non-medical images, the model can still learn relevant features that can be applied across different domains. For instance, the model might acquire knowledge about the shape and texture of kidney stones by analyzing images of rocks in mountains or stones found on beaches. The wide range and abundance of image data in ImageNet provides a valuable platform for the pre-trained model to learn transferable features that can be effectively utilized in the target model, resulting in a better model performance.
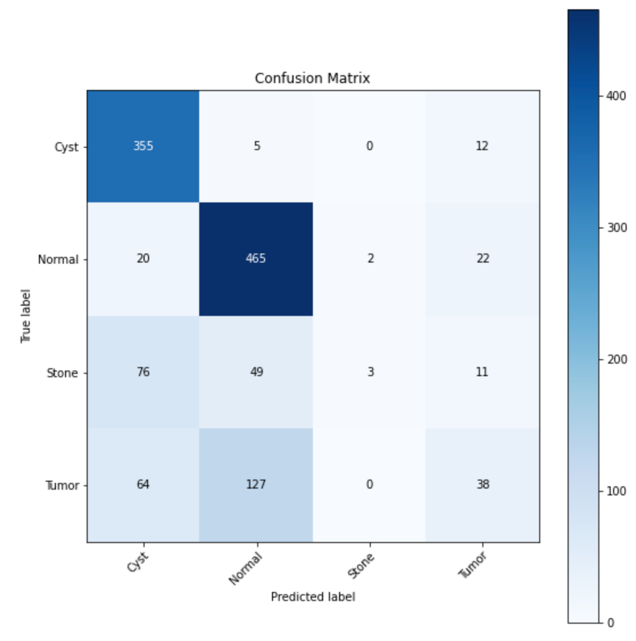
Figure 3. Confusion matrix of model without ImageNet.
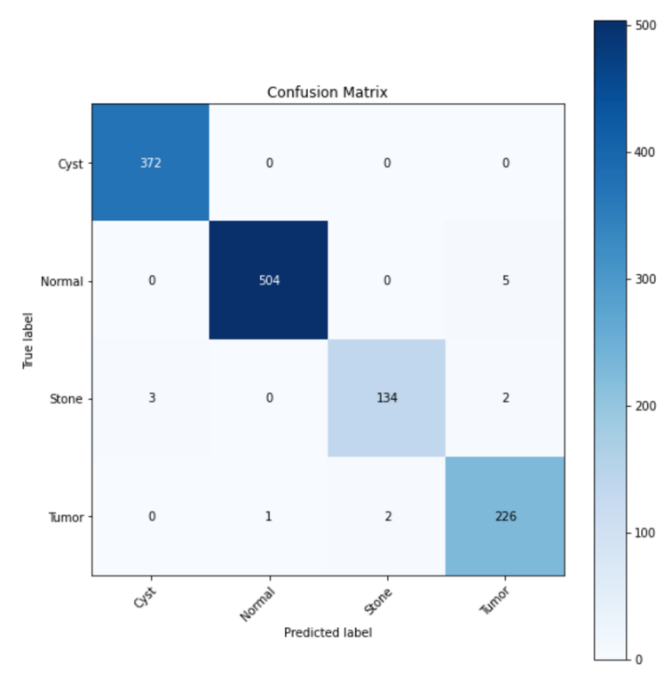
Figure 4. Confusion matrix of model with ImageNet.
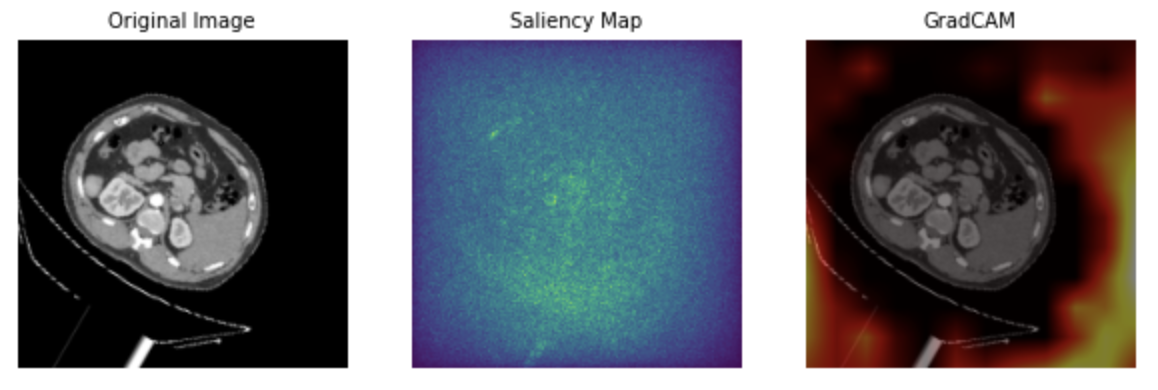
Figure 5. Sample images of model without ImageNet: original image, saliency map, and Grad-CAM.
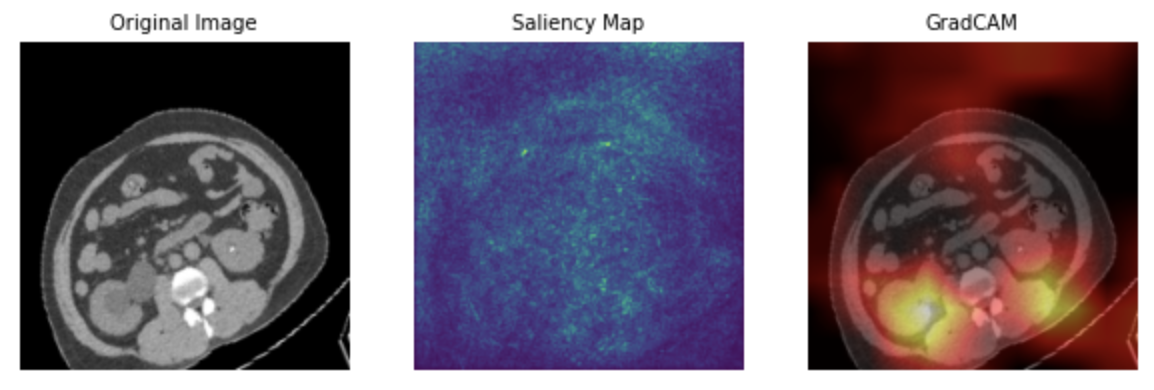
Figure 6. Sample images of model with ImageNet: original image, saliency map, and Grad-CAM.
4. Conclusion
In conclusion, this study contributes to the understanding of the effectiveness of transfer learning architectures trained on non-medical image data in the medical field. Specifically, the performance of two VGG-16 models in classifying kidney CT images was evaluated, with divergent pre-trained weights sourced from ImageNet and a random large-scale dataset. The experimental outcomes indicate that the VGG-16 model trained on ImageNet weights achieved a remarkable accuracy of 98.96%, surpassing the model trained on the random dataset by a substantial margin of approximately 30%. The validity of these results was further substantiated through the analysis of saliency maps and Grad-CAM, which demonstrated the inadequacy of the model with random pre-trained weights in accurately localizing significant features, in stark contrast to the ImageNet-trained model's proficient identification of principal features. Consequently, the utilization of the ImageNet dataset in the context of transfer learning for medical image processing was proven to be an effective approach. It is recommended that future investigations focus on refining the application of transfer learning methodologies within the medical domain to foster continued advancements in this area.
References
[1]. Reddi A S and Kuppasani K 2008 Kidney function in health and disease Nutrition and Health 3–15.
[2]. Chen T K Knicely D H and Grams M E 2019 Chronic kidney disease diagnosis and management JAMA 322 pp 1294-1304.
[3]. Bhandari M Yogarajah P Kavitha M S and Condell J 2023 Exploring the capabilities of a lightweight CNN model in accurately identifying renal abnormalities: Cysts, stones, and tumors, using lime and shap Applied Sciences 13 3125.
[4]. Islam M N Hasan M Hossain M K Alam M G Uddin M Z and Soylu A 2022 Vision transformer and explainable transfer learning models for auto detection of kidney cyst, stone and tumor from CT-radiography Scientific Reports 12 11440.
[5]. Alelign T and Petros B 2018 Kidney Stone Disease: An update on current concepts Advances in Urology 2018 pp 1–12.
[6]. Abdelrahman A and Viriri S 2022 Kidney tumor semantic segmentation using Deep Learning: A Survey of state-of-the-art Journal of Imaging 8 55.
[7]. El Nahas A M and Bello A K 2005 Chronic kidney disease: The global challenge The Lancet 365 pp 331–340.
[8]. Król E et al 2008 Early detection of chronic kidney disease: Results of the POLNEF study American Journal of Nephrology 29 pp 264–273.
[9]. Sun M Wang C Jiang F Fang X and Guo B 2019 Diagnostic value and clinical significance of ultrasound combined with CT in cystic renal cell carcinoma Oncology Letters 18(2) pp 1395-1401.
[10]. McGough W C Sanchez L E McCague C Stewart G D Schönlieb C-B Sala E and Crispin-Ortuzar M 2023 Artificial Intelligence for early detection of renal cancer in Computed Tomography: A Review Cambridge Prisms: Precision Medicine 1: E4.
[11]. Padmapriya S T and Parthasarathy S 2023 Ethical data collection for Medical Image Analysis: A Structured Approach Asian Bioethics Review pp 1-14.
[12]. Morid M A Borjali A and Del F G 2021 A scoping review of transfer learning research on medical image analysis using ImageNet Computers in Biology and Medicine 128 104115.
[13]. Xie Y Richmond D 2018 Pre-training on grayscale imagenet improves medical image classification Proceedings of the European conference on computer vision (ECCV) workshops.
[14]. Yu Q Chang C S Yan J L et al 2019 Semantic segmentation of intracranial hemorrhages in head CT scans 2019 IEEE 10th International Conference on Software Engineering and Service Science (ICSESS) IEEE pp 112-115.
[15]. Iglovikov V Shvets A 2018 Ternausnet: U-net with vgg11 encoder pre-trained on imagenet for image segmentation arXiv preprint arXiv:1801.05746.
[16]. Learning G 2021 Everything you need to know about VGG16 Medium https://medium.com/@mygreatlearning/everything-you-need-to-know-about-vgg16-7315defb5918.
[17]. Islam M N 2021 CT kidney dataset: Normal-cyst-tumor and stone Kaggle https://www.kaggle.com/datasets/nazmul0087/ct-kidney-dataset-normal-cyst-tumor-and-stone.
[18]. Yamashita R Nishio M Do R K and Togashi K 2018 Convolutional Neural Networks: An overview and application in Radiology Insights into Imaging 9 611–629.
[19]. Simonyan K and Zisserman A 2015 Very deep convolutional networks for large-scale image recognition 3rd International Conference on Learning Representations (ICLR 2015) arXiv:1409.1556.
[20]. Wei J 2019 VGG neural networks: The next step after Alexnet Medium https://towardsdatascience.com/vgg-neural-networks-the-next-step-after-alexnet-3f91fa9ffe2c.
Cite this article
Hsu,A. (2023). The investigation of transferability utilizing the ImageNet weight-based pretrained model for medical image classification: A case study on kidney CT images. Applied and Computational Engineering,22,63-70.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 5th International Conference on Computing and Data Science
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Reddi A S and Kuppasani K 2008 Kidney function in health and disease Nutrition and Health 3–15.
[2]. Chen T K Knicely D H and Grams M E 2019 Chronic kidney disease diagnosis and management JAMA 322 pp 1294-1304.
[3]. Bhandari M Yogarajah P Kavitha M S and Condell J 2023 Exploring the capabilities of a lightweight CNN model in accurately identifying renal abnormalities: Cysts, stones, and tumors, using lime and shap Applied Sciences 13 3125.
[4]. Islam M N Hasan M Hossain M K Alam M G Uddin M Z and Soylu A 2022 Vision transformer and explainable transfer learning models for auto detection of kidney cyst, stone and tumor from CT-radiography Scientific Reports 12 11440.
[5]. Alelign T and Petros B 2018 Kidney Stone Disease: An update on current concepts Advances in Urology 2018 pp 1–12.
[6]. Abdelrahman A and Viriri S 2022 Kidney tumor semantic segmentation using Deep Learning: A Survey of state-of-the-art Journal of Imaging 8 55.
[7]. El Nahas A M and Bello A K 2005 Chronic kidney disease: The global challenge The Lancet 365 pp 331–340.
[8]. Król E et al 2008 Early detection of chronic kidney disease: Results of the POLNEF study American Journal of Nephrology 29 pp 264–273.
[9]. Sun M Wang C Jiang F Fang X and Guo B 2019 Diagnostic value and clinical significance of ultrasound combined with CT in cystic renal cell carcinoma Oncology Letters 18(2) pp 1395-1401.
[10]. McGough W C Sanchez L E McCague C Stewart G D Schönlieb C-B Sala E and Crispin-Ortuzar M 2023 Artificial Intelligence for early detection of renal cancer in Computed Tomography: A Review Cambridge Prisms: Precision Medicine 1: E4.
[11]. Padmapriya S T and Parthasarathy S 2023 Ethical data collection for Medical Image Analysis: A Structured Approach Asian Bioethics Review pp 1-14.
[12]. Morid M A Borjali A and Del F G 2021 A scoping review of transfer learning research on medical image analysis using ImageNet Computers in Biology and Medicine 128 104115.
[13]. Xie Y Richmond D 2018 Pre-training on grayscale imagenet improves medical image classification Proceedings of the European conference on computer vision (ECCV) workshops.
[14]. Yu Q Chang C S Yan J L et al 2019 Semantic segmentation of intracranial hemorrhages in head CT scans 2019 IEEE 10th International Conference on Software Engineering and Service Science (ICSESS) IEEE pp 112-115.
[15]. Iglovikov V Shvets A 2018 Ternausnet: U-net with vgg11 encoder pre-trained on imagenet for image segmentation arXiv preprint arXiv:1801.05746.
[16]. Learning G 2021 Everything you need to know about VGG16 Medium https://medium.com/@mygreatlearning/everything-you-need-to-know-about-vgg16-7315defb5918.
[17]. Islam M N 2021 CT kidney dataset: Normal-cyst-tumor and stone Kaggle https://www.kaggle.com/datasets/nazmul0087/ct-kidney-dataset-normal-cyst-tumor-and-stone.
[18]. Yamashita R Nishio M Do R K and Togashi K 2018 Convolutional Neural Networks: An overview and application in Radiology Insights into Imaging 9 611–629.
[19]. Simonyan K and Zisserman A 2015 Very deep convolutional networks for large-scale image recognition 3rd International Conference on Learning Representations (ICLR 2015) arXiv:1409.1556.
[20]. Wei J 2019 VGG neural networks: The next step after Alexnet Medium https://towardsdatascience.com/vgg-neural-networks-the-next-step-after-alexnet-3f91fa9ffe2c.









