1. Introduction
Traditionally, standard medical practice has predominantly been informed by cohort-based epidemiological research, which often overlooks the nuances of individual genetic variation. Consequently, the derived conclusions are generally representative at the population level rather than the individual scale [1]. For example, dosing considerations must be tailored for distinct patient demographics, encompassing neonates, pediatric populations, geriatric individuals, patients with obesity, and those in critical care settings [2]. Therefore, the fundamental tenet of personalized medicine is predicated on the acquisition of genomics data. This concept extends beyond the patient's genome, encompassing also the genome of any pathogenic organisms, thereby facilitating a dual-faceted genomics analysis [3].
Pharmacotherapy, involving drug treatment of disease, engages with the human body through a process collectively known as pharmacokinetics, encompassing absorption, distribution, metabolism, and excretion (ADME). Initially, the body absorbs the drug, followed by its distribution and bio-transformation into active components that elicit therapeutic effects, culminating in the excretion of remaining substances. Pharmacokinetic processes are predominantly governed by specific genes, including drug metabolizing enzymes with genetic variants that influence drug processing. Concurrently, the interaction of drugs with their molecular targets, defining their pharmacodynamic properties, involves aspects such as target affinity, efficacy in modulating the target, and potency. These pharmacodynamic characteristics are also genetically regulated, determining the drug's ultimate therapeutic impact [4]. However, prior to delving into the particular gene’s role in disease manifestation, we need to take genome sequencing to know the exactly base-pairing in each gene. Luckily, recent advancements in next-generation genome sequencing have markedly decreased its cost and enhanced its throughput, thereby broadening its accessibility for research purposes [5].
After taking sequencing of genome, we can use this information to develop personalized medicine, which is an emerging, rapidly evolving approach to clinical practice where he uses new technologies to provide decision-making for the prediction, prevention, diagnosis and treatment of disease [6]. Based on identification of the patient’s genetic characteristics, personalized medicine promises to offer the precise drug at the exact dose and at the right time, making medical practice more efficient and decreasing healthcare costs [7-9].
Genomics-based pharmacokinetic (PK) modeling, a complex yet essential approach, integrates tissue physiology, anatomy, and biochemistry to forecast the tissue concentration-time relationship. It enhances understanding of metabolic enzyme kinetics, drug clearance mechanisms (including passive diffusion and transport), disposition, and excretion [8-9]. Using this method, we can optimize medical care and outcomes across diverse patient groups, thereby achieving unparalleled levels of personalized patient care [4]. In other words, genetic testing has the potential to enhance healthcare at a societal level, facilitating the administration of efficacious drugs and therapies tailored to individual patients, specific demographic groups, and broader populations [10].
However, every aspect has its pros and cons, genomics related technology is no exception. For example, as the weight of genetic information in clinical decision-making increases, patients are increasingly concerned about genetic discrimination [11]. Not only to that, the intrinsic characteristic of personalized medicine is its targeted, specific, and individualized approach, which may result in higher costs compared to traditional, broadly successful preventive interventions [12]. In this paper, we will discuss the conveniences and the difficulties in the development of personalized medicine with pharmacodynamics aspect.
2. Literature review
2.1. The development of DNA sequencing methods
The earliest DNA sequencing methods were invented and published in the 1970s by Frederick Sanger, Walter Gilbert and Allan Maxam, for example, DNA sequencing using chain termination inhibitors and DNA sequencing by chemical degradation, respectively [13-14]. In 2005, the inaugural next-generation sequencing (NGS) technology, delineated as second-generation sequencing, was conceptualized and disseminated [15-16]. This advent was swiftly followed by the introduction of a variety of NGS platforms. To surmount the challenges inherent in second-generation sequencing methods, third-generation sequencing technologies have been developed [17]. These novel technologies are characterized by their capacity for single-molecule DNA sequencing and are distinguished by enhanced throughput, augmented accuracy, extended read lengths, expedited processing times, and reduced financial implications.
2.2. The pathway of genomics sequencing development
The genome of human pathogens is being sequenced to glean insights into infection pathogenesis and potential therapeutic targets. In 1995, the inaugural complete bacterial genome of H. influenzae was sequenced [6]. Since then, over 1,000 bacterial genomes and over 3,000 viral genomes have been completely sequenced [18]. Distinct genomics loci within the reference genome are scrutinized for the identification of pathogenic variants. This process involves the utilization of multiple genomic databases, including but not limited to the 1000 Genomes Project, dbSNP, and HapMap, which aid in the filtration and annotation of variants. These variants are further evaluated based on a spectrum of defined criteria, encompassing amino acid alterations, the degree of evolutionary conservation, and the implications on protein structure. Predictive analyses of pathogenic variants leverage genetic patterns and population frequency data, as sourced from various databases (for instance, the 1000 Genomes Project), complemented by comprehensive pathway analysis [19].
This genomics data offers critical insights for comparative genomics analyses, contrasting pathogenic with non-pathogenic strains. Such analyses facilitate the identification of proteins implicated in pathogenesis, thereby presenting opportunities for the development of novel therapeutics characterized by reduced virulence [20].
3. Discussion
3.1. The advantage of personalized medicine comparing with traditional medicine
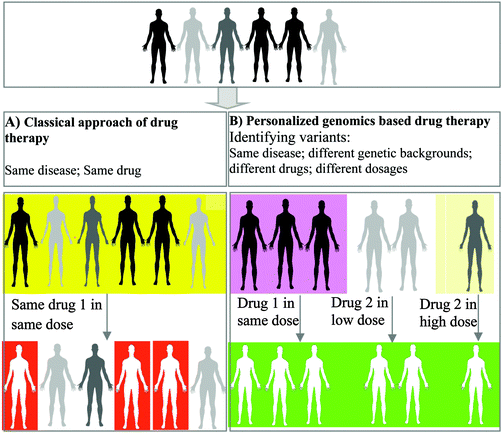
Figure 1. Traditional vs. Advanced Therapeutic Strategies in Treating Human Diseases [21]
(A) Traditional Methodology: Historically, clinicians have adopted a uniform approach to pharmacotherapy, where identical pharmaceutical agents and dosages are prescribed based on the observable phenotypic traits of a disease. This method disregards individual variability, applying a standard treatment regimen across diverse patient populations.
(B) Genomics-Driven Personalized Medicine: Contemporary advancements pivot towards a more tailored therapeutic strategy, recognizing genetic diversity as a pivotal factor in drug response. This paradigm, often termed as personalized or precision medicine, acknowledges that individuals with analogous phenotypic manifestations may require divergent pharmaceutical interventions and dosages, contingent upon their unique genetic makeup. This method employs a "genotype-first, therapy-next" strategy, which not only enhances therapeutic efficacy but also demonstrates cost-effectiveness by notably mitigating the incidence of adverse drug reactions. This approach is centered on the premise of administering the optimal drug in an appropriate dosage to the right patient, thereby revolutionizing the conventional pharmacotherapy landscape.
In conventional medical practices, treatment protocols for a specific ailment are typically uniform across all patients, derived from empirical evidence gathered from studies on populations with similar disease characteristics. This approach often applies a standardized treatment regimen to all individuals diagnosed with the same condition, irrespective of their individual differences.
3.2. Some example about personalized medicine driven by genomics
On the one hand, some illnesses are characterized by mutations and accumulation of genes occurring in a few key genes and changes in molecular paths [22]. Focusing on this illness, the personalized medicine only needs to pay attention to specific genes.
Research has increasingly focused on the impact of single-nucleotide polymorphisms (SNPs) in drug metabolism, particularly in the pharmacogenetics of immunosuppressive drugs like cyclosporine and tacrolimus [23-28]. A significant portion of inter-individual variability in the dosing of tacrolimus is attributed to polymorphisms in genes encoding proteins critical for its absorption, distribution (such as P-glycoprotein), and metabolism (CYP3A4 and CYP3A5) [29-30]. Specifically, CYP3A5 polymorphisms are closely linked to variations in blood concentrations and dose requirements for tacrolimus [31]. Studies have demonstrated that genotyping CYP3A5 can identify patients at risk for inadequate tacrolimus levels [32-33], with recipients carrying the CYP3A51 allele, from either donor or recipient, needing higher doses than those homozygous for CYP3A53 [34]. Li et al. proposed a pharmacogenetics-based dosing model incorporating CYP3A5 genotype to predict maintenance doses of tacrolimus in renal transplant recipients, enhancing initial dosing safety and efficacy [35].
However, investigations into the mechanisms underlying increased acute rejection rates have yielded mixed results [30,36]. The ABCB1 genotype of either the donor or recipient has been linked to tacrolimus distribution, acute rejection incidence, and other adverse events in kidney transplant patients [29]. But the findings regarding the impact of ABCB1 polymorphisms on the disposition and efficacy of cyclosporine and tacrolimus remain inconsistent [37, 38].
Furthermore, associations of other polymorphisms with acute rejection have been evaluated, including rs1800795 in IL-6 [39], TNF-A-308G/A [40], TLR4 rs10759932 [41], GSTM1 and GSTP1 [42], and MRP2 C24T [43]. However, due to study design limitations, these results are conflicting and do not conclusively determine the role of pharmacogenetics in shaping the individual pharmacokinetic and pharmacodynamic profiles of immunomodulators [44].
On the other hand, some illnesses are driven by multiple genes, such as stroke [9]. In this case, personalized medicine will be more complicated than single gene illness. Over the past decade or two, significant scientific progress has been made in the fields of stroke diagnosis, therapeutic interventions, and prophylactic strategies [45]. The prevailing hypothesis in contemporary medical research suggests that common stroke etiology may be attributed to the cumulative effect of multiple genes, each exerting a modest influence individually, yet collectively contributing substantially to the overall risk of stroke [46]. Similar to many disorders, individuals may possess intricate genetic profiles and distinct patterns of gene expression post-stroke, which can contribute to the risk and heightened sensitivity to ischemic stroke. In heterogeneous stroke populations, candidate gene studies address the challenge of limited patient cohorts through the deliberate selection of genes with functional relevance to specific phenotypes. This approach, often referred to as ‘association,’ is a statistical measure evaluating the dependency of a particular phenotype (such as ischemic stroke) on the presence of a specific candidate gene or allele. Consequently, associations can be positive, indicating a significant statistical relationship between the chosen gene and the phenotype [47], or negative, denoting an absence of a significant association between the gene/allele and the phenotype [47]. The selection of candidate genes is often guided by recognized stroke risk factors, including hypertension, hemostasis, and lipid metabolism abnormalities. Notably, several markers have shown significant positive associations with ischemic stroke, such as the ApoE ε2 allele and the D/D genotype of the angiotensin-converting enzyme-1 [47]. However, there are also numerous instances where genes display negative or no associations with ischemic stroke. For instance, negative associations have been identified for certain hemostasis factors (like Factor V, Q506 polymorphism, and Factor VII R353 Q polymorphism) and some hypertension factors (such as angiotensinogen and M235T polymorphism) [47].
Presently, discerning definitive patterns in candidate gene correlations with ischemic stroke presents a challenge. Moreover, the consistency of these associations across varied patient demographics, such as differing races or genetic backgrounds, remains uncertain. The complexity inherent in candidate gene association studies is further highlighted by the genomics diversity and varied phenotypic expressions within the global stroke population. Research methodologies typically employ case-control or cohort designs to closely match phenotypes among affected and unaffected individuals. Added to these variations are factors like the timing of stroke onset, environmental influences, and the degree of penetrance, meaning not all individuals with a specific genotype will exhibit the associated phenotype. Additionally, while the Human Genome Project progresses [47], the functional identification of gene products is substantially lagging, with current estimates suggesting that functions have only been assigned to about 10% of the human genome [47]. There is a significant need for further research in this domain, particularly addressing stroke genomics issues related to risk factors and the genetic underpinnings of brain vulnerability and ischemic sensitivity.
3.3. The obstacles of personalized medicine in practical application
At the theoretical application level, we need to consider many aspect in application, such as theoretical feasibility, law and local policies, cost planning, etc. First of all, we need to guarantee individual test or biomarker is accurate, reproducible and reliable. In contrast to pharmaceuticals, which are stringently regulated by entities such as the US Food and Drug Administration and various international regulatory agencies, tumor biomarker tests lack analogous oversight mechanisms. Consequently, numerous biomarker assays developed lack thorough analytical validation, casting doubt on their reliability. While some tests undergo independent analytical validation, a significant proportion do not, leading to questions regarding their dependability in clinical settings [9].
Second, a significant impediment in current clinical research is the reliance on what are termed "convenience studies." These studies, often characterized by the availability of samples or tests, generate results which may not directly address the fundamental research question. For clinicians and patients alike, it is insufficient to simply determine that a biomarker can statistically segregate a population into two distinct groups. The crucial aspect is the clinical relevance of such a division. It is imperative to understand whether the outcomes of patients can be improved by initiating or discontinuing treatment based on these biomarker results, a concept referred to as "clinical utility." At present, there exists a paucity of genomics sequencing that possess both analytical validity and clinical utility. Efforts are being made to encourage researchers to approach the development and validation of biomarker tests with the same rigor as pharmaceutical drugs, with the aim of expediting their integration into clinical practice. Such an approach is essential for establishing more robust foundational guidelines in this field [9].
Third, despite the remarkable advancements in understanding the molecular underpinnings of disease and the consequent development of therapeutics, which have notably influenced the treatment of specific cancer types, there is, to date, a lack of evidence suggesting that such progress is mirrored in the management of other complex diseases. So, if we want to promoting this model to other complex diseases may require an unimaginable amount of work [5].
On the practical level, personalized medicine can be influenced by many unexpected factors, such as education, clinical decision support, privacy, regulatory policy, standards, comparative effectiveness research, intellectual property and reimbursement [4]. A critical barrier in translating genomics information into personalized medicine is the limited knowledge of genomics among healthcare providers, coupled with restricted access to the appropriate tools. Numerous surveys indicate a growing deficit in genetic knowledge among general practitioners [48,49]. With the increasing integration of genetic data into clinical decision-making processes, there is a corresponding escalation in patient concerns regarding potential genetic discrimination [50]. If the patient worried about genetic discrimination, the personalized medicine is nowhere to talk about, that is the reason why we need a sound system to ensure patient privacy [51]. There is a requisite for policy development in the realm of regulation concerning genomics-based pharmaceutical products, particularly regarding the stringency of their testing protocols.
Not only to that, we lack a standard of personalized medicine that can be recognized by all sectors of society. The escalating oversight of genomic tools in research and clinical settings underscores the necessity for nationwide standards in genomic infrastructures. This spans from statistical methodologies to clinical trial design, and includes the establishment of a bio-banking infrastructure, which allows researchers access to data from diverse studies. Centralized and standardized systems for storing, cataloging, and annotating biological samples are integral in facilitating the progress of genomics medicine [52-54]. In this situation, in response to these challenges, Comparative Effectiveness Research (CER) has been established as a methodical approach for the evaluation of data as findings from research are integrated into clinical practice. In 2009, the American Recovery and Reinvestment Act allocated $1.1 billion specifically for CER initiatives. The Institute of Medicine delineates CER as the process of generating and synthesizing evidence to compare the benefits and detriments of various options for preventing, diagnosing, treating, and monitoring a clinical condition, or for enhancing care delivery. This approach is applicable at both individual and population levels.
4. Conclusion
The growing scrutiny and regulation of genomic tools in both research and clinical contexts highlight the critical need for unified national standards across all aspects of genomic infrastructure. This includes everything from the statistical methods used in research to the design of clinical trials, as well as the creation of comprehensive biobanking systems that provide researchers with access to a broad range of study data. The implementation of centralized and uniform frameworks for the storage, cataloging, and detailed annotation of biological samples is essential to advance the field of personalized medicine [55-56]. Although the integration of genomics research into clinical practice necessitates standardization and simplification, alongside the need to surmount barriers in education, accessibility, regulation, and reimbursement to further embed personalized medicine into clinical workflows. Genomics research has laid the groundwork for pharmacokinetics, enabling the use of patients' genomics information in making crucial clinical decisions. Personalized medicine, potentially incorporating family history and genomics data, offers a vast potential, steering healthcare towards assessing disease risk and focusing on prevention [7]. Despite current challenges in various aspects, the situation is expected to improve gradually with the development of comprehensive policies and societal oversight, leading to personalized medicine becoming a widespread treatment approach for the general populace.
References
[1]. Offit, K Personalized medicine: new genomics, old lessons. Hum. Genet. 2011, 130, 3–14.
[2]. Evans, J J Schentag, W J Jusko (Eds.), Applied Therapeutics (third ed.), Wiley Online Library (1992)
[3]. Isaac S Chan and Geoffrey S Ginsburg Annual Review of Genomics and Human Genetics 2011 12:1, 217-244
[4]. Evans, J J Schentag, W J Jusko (Eds.), Applied Therapeutics (third ed.), Wiley Online Library (1992)
[5]. Iriart J A B (2019). Precision medicine/personalized medicine: a critical analysis of movements in the transformation of biomedicine in the early 21st century. Cadernos de saúde publica, 35.
[6]. Hayes D F Markus, H S, Leslie R D et al. Personalized medicine: risk prediction, targeted therapies and mobile health technology. BMC Med 12, 37 (2014). https://doi.org/10.1186/1741-7015-12-37
[7]. European Science Foundation. Personalized medicine for the European citizen. Towards more precise medicine for the diagnosis, treatment and prevention of disease (IPM). Strasbourg: European Science Foundation; 2012.
[8]. Ghazi, I M, & Cawley, M J (2021). The science of pharmacokinetics: an overview and applications. Remington, 207-218.
[9]. Hayes D F Markus, H S, Leslie R D et al. Personalized medicine: risk prediction, targeted therapies and mobile health technology. BMC Med 12, 37 (2014). https://doi.org/10.1186/1741-7015-12-37
[10]. S Kubrick, A C Clarke, K Dullea, G Lockwood, W Sylvester, Copyright Collection (Library of Congress), et al. (1988). 2001, a space odyssey. In Criterion collection Criterion collection edit., pp. 3 videodiscs of 3 (optical) (149 min). The Voyager Company, USA.
[11]. Hall MA, McEwen JE, Barton JC, Walker AP, Howe EG, et al. 2005. Concerns in a primary care population about genetic discrimination by insurers. Genet. Med. 7: 311–16
[12]. Michael, J, Joyner, M D, & SW, R. (2015). Seven Questions for Personalized Medicine
[13]. F Sanger, S Nicklen and A R Coulson, Proc. Natl. Acad. Sci. U. S. A., 1977, 74, 5463–5467.
[14]. A M Maxam and W Gilbert, Proc. Natl. Acad. Sci. U. S. A., 1977, 74, 560–564
[15]. M Margulies, M Egholm, W E Altman, S Attiya, J S Bader and L A Bemben, et al., Nature, 2005, 437, 376–380
[16]. C Luo, D Tsementzi, N Kyrpides, T Read and K T Konstantinidis, PLoS One, 2012, 7, e30087
[17]. H Bayley, Nature, 2010, 467, 164–165
[18]. Seib KL, Dougan G, Rappuoli R. 2009. The key role of genomics in modern vaccine and drug design for emerging infectious diseases. PLoS Genet. 5: e1000612
[19]. B Rabbani, N Mahdieh, K Hosomichi, H Nakaoka and I Inoue, J Hum Genet, 2012, 57, 621–632
[20]. Rasko D A, Rosovitz M J, Myers GS, Mongodin E F, Fricke W F, et al. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 190: 6881–93
[21]. Rabbani, B, Nakaoka, H, Akhondzadeh, S, Tekin, M, & Mahdieh, N (2016). Next generation sequencing: implications in personalized medicine and pharmacogenomics. Molecular biosystems, 12(6), 1818-1830.
[22]. Di Sanzo, M, Cipolloni, L, Borro, M, La Russa, R, Santurro, A, Scopetti, M, ... & Frati, P. (2017). Clinical applications of personalized medicine: a new paradigm and challenge. Current pharmaceutical biotechnology, 18(3), 194-203.
[23]. Shuldiner, A R.; Palmer, K.; Pakyz, R E.; Alestock, T D.; Maloney, K A; O'Neill, C.; Bhatty, S; Schub, J; Overby, C L; Horenstein, R B ; Pollin, T I ; Kelemen, M D ; Beitelshees, A L; Robinson, S W; Blitzer, M G; McArdle, P F; Brown, L; Jeng, L J.; Zhao, R Y; Ambulos, N; Vesely, M R Implementation Of Pharmacogenetics: The University Of Maryland Personalized Anti-Platelet Pharmacogenetics Program. Am. J. Med. Genet. C. Semin., 2014, 166C (1), 76-84.
[24]. Beitelshees, A L; Voora, D; Lewis, J P Personalized anti-platelet and anti-coagulation therapy: applications and significance of pharmacogenomics. Pharmgenomics Pers. Med., 2015, 8, 43-61.
[25]. Pulley, J M; Denny, J C; Peterson, J F; Bernard, G R; VnencakJones, C L; Ramirez, A H; Delaney, J T; Bowton, E; Brothers, K; Johnson, K; Crawford, D C; Schildcrout, J; Masys, D R; Dilks, H H; Wilke, R A; Clayton, E W ; Shultz, E.; Laposata, M; McPherson, J; Jirjis, J N; Roden, D M Operational implementation of prospective genotyping for personalized medicine: The design of the Vanderbilt PREDICT project. Clin. Pharmacol. Ther., 2012, 92(1), 87–95.
[26]. Picard N, Marquet P The influence of pharmacogenetics and cofactors on clinical outcomes in kidney transplantation. Expert Opin Drug Metab Toxicol 2011; 7:731–43.
[27]. Haufroid V, Mourad M, Van Kerckhove V, Wawrzyniak J, De Meyer M, Eddour DC, et al. The effect of CYP3A5 and MDR1 (ABCB1) polymorphisms on cyclosporine and tacrolimus dose requirements and trough blood levels in stable renal transplant patients. Pharmacogenetics 2004; 14:147–54
[28]. Wallemacq P, Armstrong VW, Brunet M, Haufroid V, Holt DW, Johnston A, et al. Opportunities to optimize tacrolimus therapy in solid organ transplantation: report of the European consensus conference. Ther Drug Monit 2009; 31:139–52
[29]. Gómez-Bravo MA, Salcedo M, Fondevila C, Suarez F, Castellote J, Rufian S, et al. Impact of donor and recipient CYP3A5 and ABCB1 genetic polymorphisms on tacrolimus dosage requirements and rejection in Caucasian Spanish liver transplant patients. J Clin Pharmacol 2013; 53:1146–54.
[30]. Mourad M, Wallemacq P, De Meyer M, Malaise J, De Pauw L, Eddour DC, et al. Biotransformation enzymes and drug transporters pharmacogenetics in relation to immunosuppressive drugs: impact on pharmacokinetics and clinical outcome. Transplantation 2008;85: S19–24.
[31]. Hesselink DA, van Schaik RH, van Agteren M, de Fijter JW, Hartmann A, Zeier M, et al. CYP3A5 genotype is not associated with a higher risk of acute rejection in tacrolimus-treated renal transplant recipients. Pharmacogenet Genomics 2008; 18:339–48.
[32]. Thervet E, Loriot MA, Barbier S, Buchler M, Ficheux M, Choukroun G, et al. Optimization of initial tacrolimus dose using pharmacogenetic testing. Clin Pharmacol Ther 2010; 87:721–6.
[33]. Van Gelder T, Hesselink DA. Dosing tacrolimus based on CYP3A5 genotype: will it improve clinical outcome? Clin Pharmacol Ther 2010; 87:640–1.
[34]. Quteineh L, Verstuyft C Pharmacogenetics in immunosuppressants: impact on dose requirement of calcineurin inhibitors in renal and liver pediatric transplant recipients. Curr Opin Organ Transplant 2010; 15:601–7.
[35]. Li L, Li C-J, Zheng L, Zhang Y-J, Jiang H-X, Si-Tu B, et al. Tacrolimus dosing in Chinese renal transplant recipients: a population-based pharmacogenetics study. Eur J Clin Pharmacol 2011; 67:787–95.
[36]. Rahsaz M, Azarpira N, Nikeghbalian S, Aghdaie MH, Geramizadeh B, Moini M, et al. Association between tacrolimus concentration and genetic polymorphisms of CYP3A5 and ABCB1 during the early stage after liver transplant in an Iranian population. Exp Clin Transplant 2012; 10:24–9.
[37]. Hauser I A, Schaeffeler E, Gauer S, Scheuermann EH, Wegner B, Gossmann J, et al. ABCB1 genotype of the donor but not of the recipient is a major risk factor for cyclosporine-related nephrotoxicity after renal transplantation. J Am Soc Nephrol 2005; 16:1501–11.
[38]. Woillard J-B, Rerolle J-P, Picard N, Rousseau A, Guillaudeau A, Munteanu E, et al. Donor P-gp polymorphisms strongly influence renal function and graft loss in a cohort of renal transplant recipients on cyclosporine therapy in a long-term follow-up. Clin Pharmacol Ther 2010; 88:95–100.
[39]. Lv R, Hu X, Bai Y, Long H, Xu L, Liu Z, et al. Association between IL-6-174G/C polymorphism and acute rejection of renal allograft: evidence from a meta-analysis. Transpl Immunol 2012; 26:11–8.
[40]. Hu X, Bai Y, Li S, Zeng K, Xu L, Liu Z, et al. Donor or recipient TNF-A-308G/A polymorphism and acute rejection of renal allograft: a meta-analysis. Transpl Immunol 2011; 25:61–71.
[41]. Hwang Y-H, Ro H, Choi I, Kim H, Oh K-H, Hwang J-I, et al. Impact of polymorphisms of TLR4/CD14 and TLR3 on acute rejection in kidney transplantation. Transplantation 2009; 88:699–705
[42]. Singh R, Manchanda PK, Kesarwani P, Srivastava A, Mittal RD. Influence of genetic polymorphisms in GSTM1, GSTM3, GSTT1 and GSTP1 on allograft outcome in renal transplant recipients. Clin Transplant 2009; 23:490–8.
[43]. Lloberas N, Torras J, Cruzado JM, Andreu F, Oppenheimer F, Sánchez-Plumed J, et al. Influence of MRP2 on MPA pharmacokinetics in renal transplant recipients-results of the Pharmacogenomic Substudy within the Symphony Study. Nephrol Dial Transplant 2011; 26:3784–93.
[44]. García-González, X, Cabaleiro, T, Herrero, M, McLeod, H & López-Fernández, L. (2016). Clinical implementation of pharmacogenetics. Drug Metabolism and Personalized Therapy, 31(1), 9-16.
[45]. Donnan G.A., Fisher M., Macleod M. and Davis S.M.: "Stroke". Lancet 2008; 371: 1612.
[46]. Hunter D J, Altshuler D and Rader D J: "From Darwin’s finches to canaries in the coal mine—mining the genome for new biology". N Engl J Med 2008; 358: 2760
[47]. Read SJ, Parsons AA, Harrison D C, et al. Stroke Genomics: Approaches to Identify, Validate, and Understand Ischemic Stroke Gene Expression. Journal of Cerebral Blood Flow & Metabolism. 2001;21(7):755-778. doi:10.1097/00004647-200107000-00001
[48]. Lapham E V, Kozma C, Weiss J O, Benkendorf JL, Wilson M A. 2000. The gap between practice and genetics education of health professionals: HuGEM survey results. Genet. Med. 2: 226–31
[49]. Metcalfe S, Hurworth R, Newstead J, Robins R. 2002. Needs assessment study of genetics education for general practitioners in Australia. Genet. Med. 4: 71–77
[50]. Hall MA, McEwen J E, Barton J C, Walker A P, Howe E G, et al. 2005. Concerns in a primary care population about genetic discrimination by insurers. Genet. Med. 7: 311–16
[51]. Apse KA, Biesecker B B, Giardiello F M, Fuller B P, Bernhardt BA. 2004. Perceptions of genetic discrimination among at-risk relatives of colorectal cancer patients. Genet. Med. 6: 510–16
[52]. Vastag B. 2006. New clinical trials policy at FDA. Nat. Biotechnol. 24: 1043
[53]. Ginsburg GS, Burke T W, Febbo P. 2008. Centralized biorepositories for genetic and genomic research. JAMA 299: 1359–61
[54]. Westfall J M, Mold J, Fagnan L. 2007. Practice-based research: “Blue Highways” on the NIH roadmap. JAMA 297: 403–6
[55]. Offit, K Personalized medicine: new genomics, old lessons. Hum. Genet. 2011, 130, 3–14.
[56]. Inst. Med. Comm. Comp. Eff. Res. Prioritization. 2009. Initial National Priorities for Comparative Effectiveness Research. Washington, DC: National Academies. 227 pp.
Cite this article
Shao,Y. (2024). Genomics-driven pharmacodynamics: A new frontier in personalized medicine. Theoretical and Natural Science,35,164-172.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 2nd International Conference on Modern Medicine and Global Health
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Offit, K Personalized medicine: new genomics, old lessons. Hum. Genet. 2011, 130, 3–14.
[2]. Evans, J J Schentag, W J Jusko (Eds.), Applied Therapeutics (third ed.), Wiley Online Library (1992)
[3]. Isaac S Chan and Geoffrey S Ginsburg Annual Review of Genomics and Human Genetics 2011 12:1, 217-244
[4]. Evans, J J Schentag, W J Jusko (Eds.), Applied Therapeutics (third ed.), Wiley Online Library (1992)
[5]. Iriart J A B (2019). Precision medicine/personalized medicine: a critical analysis of movements in the transformation of biomedicine in the early 21st century. Cadernos de saúde publica, 35.
[6]. Hayes D F Markus, H S, Leslie R D et al. Personalized medicine: risk prediction, targeted therapies and mobile health technology. BMC Med 12, 37 (2014). https://doi.org/10.1186/1741-7015-12-37
[7]. European Science Foundation. Personalized medicine for the European citizen. Towards more precise medicine for the diagnosis, treatment and prevention of disease (IPM). Strasbourg: European Science Foundation; 2012.
[8]. Ghazi, I M, & Cawley, M J (2021). The science of pharmacokinetics: an overview and applications. Remington, 207-218.
[9]. Hayes D F Markus, H S, Leslie R D et al. Personalized medicine: risk prediction, targeted therapies and mobile health technology. BMC Med 12, 37 (2014). https://doi.org/10.1186/1741-7015-12-37
[10]. S Kubrick, A C Clarke, K Dullea, G Lockwood, W Sylvester, Copyright Collection (Library of Congress), et al. (1988). 2001, a space odyssey. In Criterion collection Criterion collection edit., pp. 3 videodiscs of 3 (optical) (149 min). The Voyager Company, USA.
[11]. Hall MA, McEwen JE, Barton JC, Walker AP, Howe EG, et al. 2005. Concerns in a primary care population about genetic discrimination by insurers. Genet. Med. 7: 311–16
[12]. Michael, J, Joyner, M D, & SW, R. (2015). Seven Questions for Personalized Medicine
[13]. F Sanger, S Nicklen and A R Coulson, Proc. Natl. Acad. Sci. U. S. A., 1977, 74, 5463–5467.
[14]. A M Maxam and W Gilbert, Proc. Natl. Acad. Sci. U. S. A., 1977, 74, 560–564
[15]. M Margulies, M Egholm, W E Altman, S Attiya, J S Bader and L A Bemben, et al., Nature, 2005, 437, 376–380
[16]. C Luo, D Tsementzi, N Kyrpides, T Read and K T Konstantinidis, PLoS One, 2012, 7, e30087
[17]. H Bayley, Nature, 2010, 467, 164–165
[18]. Seib KL, Dougan G, Rappuoli R. 2009. The key role of genomics in modern vaccine and drug design for emerging infectious diseases. PLoS Genet. 5: e1000612
[19]. B Rabbani, N Mahdieh, K Hosomichi, H Nakaoka and I Inoue, J Hum Genet, 2012, 57, 621–632
[20]. Rasko D A, Rosovitz M J, Myers GS, Mongodin E F, Fricke W F, et al. 2008. The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J. Bacteriol. 190: 6881–93
[21]. Rabbani, B, Nakaoka, H, Akhondzadeh, S, Tekin, M, & Mahdieh, N (2016). Next generation sequencing: implications in personalized medicine and pharmacogenomics. Molecular biosystems, 12(6), 1818-1830.
[22]. Di Sanzo, M, Cipolloni, L, Borro, M, La Russa, R, Santurro, A, Scopetti, M, ... & Frati, P. (2017). Clinical applications of personalized medicine: a new paradigm and challenge. Current pharmaceutical biotechnology, 18(3), 194-203.
[23]. Shuldiner, A R.; Palmer, K.; Pakyz, R E.; Alestock, T D.; Maloney, K A; O'Neill, C.; Bhatty, S; Schub, J; Overby, C L; Horenstein, R B ; Pollin, T I ; Kelemen, M D ; Beitelshees, A L; Robinson, S W; Blitzer, M G; McArdle, P F; Brown, L; Jeng, L J.; Zhao, R Y; Ambulos, N; Vesely, M R Implementation Of Pharmacogenetics: The University Of Maryland Personalized Anti-Platelet Pharmacogenetics Program. Am. J. Med. Genet. C. Semin., 2014, 166C (1), 76-84.
[24]. Beitelshees, A L; Voora, D; Lewis, J P Personalized anti-platelet and anti-coagulation therapy: applications and significance of pharmacogenomics. Pharmgenomics Pers. Med., 2015, 8, 43-61.
[25]. Pulley, J M; Denny, J C; Peterson, J F; Bernard, G R; VnencakJones, C L; Ramirez, A H; Delaney, J T; Bowton, E; Brothers, K; Johnson, K; Crawford, D C; Schildcrout, J; Masys, D R; Dilks, H H; Wilke, R A; Clayton, E W ; Shultz, E.; Laposata, M; McPherson, J; Jirjis, J N; Roden, D M Operational implementation of prospective genotyping for personalized medicine: The design of the Vanderbilt PREDICT project. Clin. Pharmacol. Ther., 2012, 92(1), 87–95.
[26]. Picard N, Marquet P The influence of pharmacogenetics and cofactors on clinical outcomes in kidney transplantation. Expert Opin Drug Metab Toxicol 2011; 7:731–43.
[27]. Haufroid V, Mourad M, Van Kerckhove V, Wawrzyniak J, De Meyer M, Eddour DC, et al. The effect of CYP3A5 and MDR1 (ABCB1) polymorphisms on cyclosporine and tacrolimus dose requirements and trough blood levels in stable renal transplant patients. Pharmacogenetics 2004; 14:147–54
[28]. Wallemacq P, Armstrong VW, Brunet M, Haufroid V, Holt DW, Johnston A, et al. Opportunities to optimize tacrolimus therapy in solid organ transplantation: report of the European consensus conference. Ther Drug Monit 2009; 31:139–52
[29]. Gómez-Bravo MA, Salcedo M, Fondevila C, Suarez F, Castellote J, Rufian S, et al. Impact of donor and recipient CYP3A5 and ABCB1 genetic polymorphisms on tacrolimus dosage requirements and rejection in Caucasian Spanish liver transplant patients. J Clin Pharmacol 2013; 53:1146–54.
[30]. Mourad M, Wallemacq P, De Meyer M, Malaise J, De Pauw L, Eddour DC, et al. Biotransformation enzymes and drug transporters pharmacogenetics in relation to immunosuppressive drugs: impact on pharmacokinetics and clinical outcome. Transplantation 2008;85: S19–24.
[31]. Hesselink DA, van Schaik RH, van Agteren M, de Fijter JW, Hartmann A, Zeier M, et al. CYP3A5 genotype is not associated with a higher risk of acute rejection in tacrolimus-treated renal transplant recipients. Pharmacogenet Genomics 2008; 18:339–48.
[32]. Thervet E, Loriot MA, Barbier S, Buchler M, Ficheux M, Choukroun G, et al. Optimization of initial tacrolimus dose using pharmacogenetic testing. Clin Pharmacol Ther 2010; 87:721–6.
[33]. Van Gelder T, Hesselink DA. Dosing tacrolimus based on CYP3A5 genotype: will it improve clinical outcome? Clin Pharmacol Ther 2010; 87:640–1.
[34]. Quteineh L, Verstuyft C Pharmacogenetics in immunosuppressants: impact on dose requirement of calcineurin inhibitors in renal and liver pediatric transplant recipients. Curr Opin Organ Transplant 2010; 15:601–7.
[35]. Li L, Li C-J, Zheng L, Zhang Y-J, Jiang H-X, Si-Tu B, et al. Tacrolimus dosing in Chinese renal transplant recipients: a population-based pharmacogenetics study. Eur J Clin Pharmacol 2011; 67:787–95.
[36]. Rahsaz M, Azarpira N, Nikeghbalian S, Aghdaie MH, Geramizadeh B, Moini M, et al. Association between tacrolimus concentration and genetic polymorphisms of CYP3A5 and ABCB1 during the early stage after liver transplant in an Iranian population. Exp Clin Transplant 2012; 10:24–9.
[37]. Hauser I A, Schaeffeler E, Gauer S, Scheuermann EH, Wegner B, Gossmann J, et al. ABCB1 genotype of the donor but not of the recipient is a major risk factor for cyclosporine-related nephrotoxicity after renal transplantation. J Am Soc Nephrol 2005; 16:1501–11.
[38]. Woillard J-B, Rerolle J-P, Picard N, Rousseau A, Guillaudeau A, Munteanu E, et al. Donor P-gp polymorphisms strongly influence renal function and graft loss in a cohort of renal transplant recipients on cyclosporine therapy in a long-term follow-up. Clin Pharmacol Ther 2010; 88:95–100.
[39]. Lv R, Hu X, Bai Y, Long H, Xu L, Liu Z, et al. Association between IL-6-174G/C polymorphism and acute rejection of renal allograft: evidence from a meta-analysis. Transpl Immunol 2012; 26:11–8.
[40]. Hu X, Bai Y, Li S, Zeng K, Xu L, Liu Z, et al. Donor or recipient TNF-A-308G/A polymorphism and acute rejection of renal allograft: a meta-analysis. Transpl Immunol 2011; 25:61–71.
[41]. Hwang Y-H, Ro H, Choi I, Kim H, Oh K-H, Hwang J-I, et al. Impact of polymorphisms of TLR4/CD14 and TLR3 on acute rejection in kidney transplantation. Transplantation 2009; 88:699–705
[42]. Singh R, Manchanda PK, Kesarwani P, Srivastava A, Mittal RD. Influence of genetic polymorphisms in GSTM1, GSTM3, GSTT1 and GSTP1 on allograft outcome in renal transplant recipients. Clin Transplant 2009; 23:490–8.
[43]. Lloberas N, Torras J, Cruzado JM, Andreu F, Oppenheimer F, Sánchez-Plumed J, et al. Influence of MRP2 on MPA pharmacokinetics in renal transplant recipients-results of the Pharmacogenomic Substudy within the Symphony Study. Nephrol Dial Transplant 2011; 26:3784–93.
[44]. García-González, X, Cabaleiro, T, Herrero, M, McLeod, H & López-Fernández, L. (2016). Clinical implementation of pharmacogenetics. Drug Metabolism and Personalized Therapy, 31(1), 9-16.
[45]. Donnan G.A., Fisher M., Macleod M. and Davis S.M.: "Stroke". Lancet 2008; 371: 1612.
[46]. Hunter D J, Altshuler D and Rader D J: "From Darwin’s finches to canaries in the coal mine—mining the genome for new biology". N Engl J Med 2008; 358: 2760
[47]. Read SJ, Parsons AA, Harrison D C, et al. Stroke Genomics: Approaches to Identify, Validate, and Understand Ischemic Stroke Gene Expression. Journal of Cerebral Blood Flow & Metabolism. 2001;21(7):755-778. doi:10.1097/00004647-200107000-00001
[48]. Lapham E V, Kozma C, Weiss J O, Benkendorf JL, Wilson M A. 2000. The gap between practice and genetics education of health professionals: HuGEM survey results. Genet. Med. 2: 226–31
[49]. Metcalfe S, Hurworth R, Newstead J, Robins R. 2002. Needs assessment study of genetics education for general practitioners in Australia. Genet. Med. 4: 71–77
[50]. Hall MA, McEwen J E, Barton J C, Walker A P, Howe E G, et al. 2005. Concerns in a primary care population about genetic discrimination by insurers. Genet. Med. 7: 311–16
[51]. Apse KA, Biesecker B B, Giardiello F M, Fuller B P, Bernhardt BA. 2004. Perceptions of genetic discrimination among at-risk relatives of colorectal cancer patients. Genet. Med. 6: 510–16
[52]. Vastag B. 2006. New clinical trials policy at FDA. Nat. Biotechnol. 24: 1043
[53]. Ginsburg GS, Burke T W, Febbo P. 2008. Centralized biorepositories for genetic and genomic research. JAMA 299: 1359–61
[54]. Westfall J M, Mold J, Fagnan L. 2007. Practice-based research: “Blue Highways” on the NIH roadmap. JAMA 297: 403–6
[55]. Offit, K Personalized medicine: new genomics, old lessons. Hum. Genet. 2011, 130, 3–14.
[56]. Inst. Med. Comm. Comp. Eff. Res. Prioritization. 2009. Initial National Priorities for Comparative Effectiveness Research. Washington, DC: National Academies. 227 pp.