1. Introduction
Western Blotting (WB) and quantitative Polymerase Chain Reaction (qPCR) are widely regarded as the gold standard detection techniques in molecular biology, which play pivotal roles in the separation, visualization, identification, and quantification of biomacromolecules [1, 2]. These two methodologies detect different analytes: WB specifically focuses on proteins, whereas qPCR primarily aims to detect and quantify DNA or RNA.Despite their extensive application, both WB and qPCR technologies possess inherent limitations. Hence, enhancing these traditional techniques is a prevalent trend in current research endeavors. The advent of novel technologies heralds the gradual supplanting of conventional methods and promises to propel advancements in medical research and healthcare delivery.
2. Bottlenecks and limitations of traditional WB and qPCR
2.1. Western Bloting
Western Blotting is a widely employed method for detecting post-translational modifications and elucidating protein structures [3], traditionally considered a semi-quantitative technique. This method identifies one or more protein targets through a foundational step and two pivotal procedures: protein extraction, SDS-PAGE separation, and immunoblotting. It encompasses three critical components: molecular weight-based separation, transfer onto an adsorptive membrane, and identification of target proteins using specific primary and secondary antibodies [4]. Protein extraction is the cornerstone of Western Blot experiments. It involves lysing cells or tissues to release proteins. This process is usually facilitated by an appropriate lysis buffer at low temperatures to maintain protein integrity. SDS-PAGE electrophoresis separates proteins according to their molecular weight based on their migration patterns in an electric field, necessitating meticulous sample preparation and the establishment of suitable electrophoresis conditions for effective resolution. The transmembrane transfer step is crucial, where proteins are shifted from the gel onto nitrocellulose or polyvinylidene fluoride membranes [5], often employing wet or semi-dry transfer methods [6]. Care must be taken with transfer duration and voltage to guarantee efficient protein transfer. Following transfer, specific antibodies are incubated with the membrane-bound proteins to target the protein of interest, with imaging converting these interactions into visible bands, thereby enabling the detection of target protein expression levels. Results are interpreted by comparing the relative abundance of target proteins with internal reference proteins
Despite its prevalent use in laboratories, traditional Western Blot technology possesses notable limitations.
Semi-quantitative Nature: Traditional Western Blotting is inherently semi-quantitative. When comparing across sample groups or different experiments, precise quantification of protein expression levels is unattainable [7]. Establishing a linear relationship between measurable outcomes and protein mass is essential, influenced by factors such as the blotting protocol, antibody binding kinetics, chemiluminescent reactions, and image acquisition and processing techniques [8]. In proteomics, techniques like mass spectrometry offer more comprehensive quantitative solutions [9], while non-blotting technologies introduce a novel quality control tool for data standardization in Western Blot workflows. By assessing the optical density of total protein bands, the ratio of target protein band optical density to total protein band optical density can be utilized for target protein quantification [10].
Limited Linear Range: The narrow linear range restricts its effectiveness in detecting low-abundance proteins. For instance, the commonly utilized reference protein β-actin exhibits a limited linear dynamic range of only 3.75-15μg in quantitative Western Blot analysis [11].
Sensitivity Constraints: Traditional Western Blotting often fails to detect low-abundance target proteins, such as EGFR, due to insufficient sensitivity to capture these subtle signals. While theoretically, higher protein expression should correspond to greater grayscale values, practical grayscale values may deviate from theoretical expectations, potentially due to signal saturation or other experimental variables [12].
Low Throughput: Traditional Western Blotting can only assess one or a few proteins simultaneously, which is inefficient and impractical for large-scale proteomics research endeavors.
2.2. QPCR
qPCR, or real-time quantitative Polymerase Chain Reaction, has the ability to amplify specific sequences of small amounts of Deoxyribonucleic Acid (DNA) molecules to detectable levels [13]. By incorporating a fluorescent group, it enables real-time monitoring of the PCR process, akin to installing a "real-time monitor" for the PCR reaction, allowing for dynamic observation of the entire DNA amplification process. The primary distinction between qPCR and traditional PCR lies in its capacity to not only qualitatively detect the presence of DNA but also accurately quantify the initial template, providing more precise and comprehensive data [14] for research and clinical diagnosis.
While qPCR represents a significant improvement over traditional PCR technology in terms of quantitative analysis, it still faces several limitations.
Interference from Non-specific Products in Dye-based qPCR: In dye-based qPCR, SYBR Green I binds to any double-stranded DNA, such as primer dimers. This binding increases fluorescence signals, interfering with quantitative accuracy [15]. For instance, when detecting antibiotic resistance genes, the signal from primer dimers may overshadow the target gene signal, resulting in false positives [16]. Although probe-based methods offer high specificity, probe design must consider factors such as sequence specificity and secondary structure, making them complex and costly, thereby limiting their widespread application. This challenge is particularly pronounced in multiplex gene detection, where multiple specific probes need to be designed for detecting multiple genes, and interference between probes must be avoided, increasing experimental design complexity and costs [17].
Sensitivity and Quantitative Limitations: The quantitative results of qPCR are significantly influenced by various factors, including RNA templates, assay design variability, and issues with data normalization and analysis consistency. When gene copy numbers are compared across different laboratories, the results are often difficult to directly compare due to variations in standards, hindering research and clinical applications. Moreover, the detection sensitivity of SYBR Green-based qPCR is limited, making it challenging to quantify the expression of low-abundance genes or RNA samples extracted from highly restricted or limited sources [18]. For example, when detecting circulating tumor DNA in blood, its concentration can be extremely low [19], and traditional PCR may fail to detect it accurately, posing a risk of missed diagnosis.
Multiple Detection and Throughput Bottleneck: Traditional qPCR typically operates at low to medium throughput, as most qPCR platforms utilize a 96- or 384-well plate format [20,21]. This may not suffice for the simultaneous detection of multiple genes. When detecting multiple gene mutations, it necessitates multiple reactions, escalating both experimental costs and time. During flu seasons, when conducting pathogen testing on a large number of patient samples, traditional PCR struggles to complete the task expeditiously.
With the swift advancements in biotechnology, the limitations of traditional WB and qPCR techniques in gene and protein detection have become increasingly evident. These methods often grapple with low throughput, semi-quantitative results, and low sensitivity, failing to meet the demands for efficient and rapid testing, especially in disease screening and drug diagnosis, where they exhibit significant shortcomings. However, emerging WB and qPCR technologies are transforming this landscape, offering faster and more efficient means for gene and protein detection, showcasing their substantial advantages in medical diagnostics.
3. Emerging WB technology
Traditional WB technology has limitations such as low throughput, semi-quantitative and low sensitivity, while the emerging WB technology is gradually solving these problems. Common new WB technologies include spatial resolution WB, high sensitivity WB, high throughput WB, etc., each of which has unique advantages.
3.1. Spatial resolution WB
This method arranges different tumor tissue samples on a slide according to certain rules and makes them into tissue chips. Multiple samples are analyzed simultaneously on a chip to improve detection efficiency and facilitate the comparison of differences between different samples.
3.1.1. Single-cell resolution WB
Single-cell resolution WB involves using microfabricated devices to perform WB analysis on individual cells, measuring differences in protein expression levels and states between cells.In Figure 1,the process mainly consists of five stages: gravitational sedimentation of cells into microwells; chemical lysis of cells in each micwell; PAGE of each single-cell lysate; exposing the gel to UV light to blot (fix) proteins onto the gel matrix; and in situ immunodetection [22] of fixed proteins within the gel. Its innovation primarily lies in the use of UV light to fix proteins and the multiplex detection capabilities (Figure 1(a)).
ScWB fixes separated proteins onto Polyacrylamide Gel (PAG) via ultraviolet light irradiation. This maintains a high local protein concentration for in-gel immunodetection [23] and avoids the loss and inhomogeneity caused by the transfer step in traditional WB. By using antibody mixtures for detection and antibody stripping/re-detection techniques, it is possible to detect over 10 proteins in each cell, breaking the limitations of traditional low-throughput WB. Once micro-devices are fabricated, researchers can complete the detection within 4-6 hours and generate highly selective multiplex data from individual cells.
Single-cell resolution Western Blotting is primarily utilized in cancer cell research and in the screening and evaluation of drugs.
Single-cell spatially resolved WB can be used to analyze the differential responses of cancer cells to different stimuli or drugs, revealing heterogeneity among individual cancer cells; it can also detect the expression levels and modification states of specific proteins in cancer cells, thereby aiding in understanding tumor progression, drug resistance mechanisms, and the design of personalized treatment strategies. Studies have used imaging and scWestern analysis on individual glioblastoma cells treated with the chemotherapeutic agent daunorubicin to identify apoptotic and surviving cells. The surviving glioblastoma subpopulation showed upregulated expression of the multidrug resistance protein P-glycoprotein(P-gp), suggesting that active drug efthroughput pumps may be a potential mechanism for drug resistance.
scWB can measure changes in key proteins during the differentiation of individual stem cells, identifying markers at various stages of differentiation, providing crucial information for stem cell biology and regenerative medicine. By imaging and performing single-cell Western Blot analysis on individual glioblastoma cells treated with chemotherapeutic drugs, it was found that among those undergoing specific differentiation stages (such as markers of apoptosis cleaved caspase 8 and annexin V), there are both apoptotic cells (positive for cleaved caspase 8 and annexinV) and surviving cells [24].
Although single-cell immunoassay methods are powerful, the throughput and sensitivity of detection remain analytical challenges [25].
The single-experiment throughput of spatial-resolution WB technology is relatively low. Each micro-device can process about 10^3 cells.This is mainly due to the trade-off between the size of standard microscope slides and the surface area required for each single-cell experiment. While increasing the separation distance can boost throughput, this would reduce peak capacity. On standard microscope slides, the surface area required for each single-cell experiment is about 0.4×1 mm. This size limitation imposes certain limitations on the handling of large cell samples in spatially resolved WB technology.
In addition, the detection sensitivity of this technology is relatively low, with a minimum detection limit of approximately 27,000 protein molecules per cell, which indicates that the technology can detect only the top 50% of proteins present in mammalian proteomes [26]. For detecting low-abundance proteins, it is necessary to further improve detection sensitivity. For instance, in the study of certain low-abundance proteins, spatially resolved Western Blot (WB) techniques may not detect their expression. This limitation restricts the application of this technology in researching the functions and mechanisms of low-abundance proteins.
3.1.2. Subcellular resolution WB ((sc)2WB)
In cell populations, cellular properties may exhibit significant differences [27], including but not limited to changes in cell state, diversity in gene expression patterns, and differences in protein function. The subcellular localization of proteins is closely related to their function.
Subcellular resolution WB technology has developed a pair of orthogonal lysis buffers that can separately lyse the cytoplasm and nucleus, and use them as electrophoresis buffers; a bidirectional PAGE analysis method was designed and validated for independently measuring and spatially separating target protein expression in the cytoplasm and nuclear compartments, which differs slightly from single-cell resolution WB.
This technology not only enhances the selectivity of detection but also significantly boosts throughput, providing a powerful tool for in-depth research on cellular heterogeneity and protein localization. NF-κB is a pivotal regulatory factor in numerous diseases, including cancer and autoimmune disorders [28], and changes in its dynamic distribution may indicate disease progression or treatment efficacy. (sc)2WB can measure the dynamic translocation of NF-κB from the cytoplasm to the nucleus at intervals of 15 minutes. This capability is crucial for understanding signaling mechanisms in inflammation, immune responses, and cancers.
Although subcellular resolution WB technology has significant advantages in reducing technical variation, improving resolution, and increasing throughput, it still has some limitations. These include insufficient optimization of lysis buffer, short lysis time constraints, limited multiplexing capabilities, challenges in scaling up throughput, and room for improvement in subcellular resolution. These issues point the way forward for future research and technological advancements.
To sum up, space resolution WB technology currently has the demand of high throughput and sensitivity. The following is mainly a technical exploration for high throughput and high sensitivity.
3.2. High sensitivity WB
In view of the insufficient sensitivity in traditional WB and spatial resolution WB, researchers have developed relevant technologies to increase the sensitivity of WB detection.
3.2.1. Nanoblotting technique
Dot blotting improves on traditional Western Blot and is a validated technique used as an alternative to protein separation processes. Traditional dot blotting involves spotting protein samples on membrane paper. Then, proteins are identified and quantified through immunoassay procedures, like colorimetry, fluorescence, or chemiluminescence. However, it requires a large amount of sample and antibody (50 to 100μL) to detect the target protein [29]. In contrast, alternative nano-dot blotting has lower sample requirements, simplified experimental steps, high-throughput analysis, and cost-effectiveness. This method uses a self-made multi-nozzle device to deposit approximately 200nL of sample onto nitrocellulose membranes, thereby reducing the need for both sample and antibody. Although the tubing needs to be filled before the spotting procedure, this method can easily prepare about 50 samples based on the required sample volume for each spot [30].
Dot blot can be used in the detection of G Protein-Coupled Receptors (GPCRs). GPCRs constitute the largest family of membrane proteins and have significant pharmacological importance, as approximately 50% of drugs on the market exert their effects through interactions with G protein-coupled receptors [31]. Dot blot technology is used to detect the expression levels and activity of G protein-coupled receptors, thereby studying the pharmacological effects of drugs. Nano-dot blot has also been applied in rat models of acute and chronic epileptic seizures to detect six proteins associated with inflammatory processes. This technique is still under development and has room for improvement.
3.2.2. Quantum dot labeling technology
The method of detecting and counting discrete Quantum Dots (QDs) in transparent PVDF films is called Single Point Quantum Dot (SPQD) protein blot [32].
QD-labeling technology improves the Western Blot detection method. It significantly enhances the performance of trace protein detection by counting fluorescent spots on an optically transparent PVDF membrane. Bakalova et al. replaced traditional HRP chemiluminescent labeling with bright quantum dots, markedly improving the sensitivity of Western Blot detection [33]. This method achieves optimal detection sensitivity at 0.2pg and requires a minimal sample size of 100 cells, which is 100 times more sensitive and 100 times less cell usage compared to traditional Western Blot.
CdTe is a multifunctional semiconductor material with extensive application potential in optoelectronics and nuclear detection [34]. When CdTe quantum dots are coated with biotinylated denatured bovine serum albumin, the resulting QDs are strongly fluorescent. These QDs are linked together via the biotin-avidin system The linked quantum dots are called POLY-QDs, which serve as a classic marker in quantum dot labeling techniques. Through this series of modifications, the fluorescence intensity of CdTe-QDs is significantly increased, making them more sensitive than traditionally stained WB [35].
Quantum Dots (QDs), widely used in chemistry and biology because of their special optical and electronic properties, can label neuronal receptors, activate downstream signaling pathways, and aid in the study of neuronal growth and signal transduction mechanisms. Quantum dots can label neuronal receptors and activate downstream signaling pathways, aiding in the study of neuronal growth and signal transduction mechanisms. NGF is a hormone protein [36] associated with many pathophysiological conditions, and SPQD has been used to detect in purified nerve growth factor using polyclonal anti-NGF antibodies.
The high brightness and stability of quantum dots make them ideal probes for in vivo imaging, suitable for real-time monitoring of dynamic changes in intracellular molecules, cell labeling, tissue imaging, and in vivo animal imaging. Some studies have utilized antibody-conjugated quantum dots to detect specific pathogens (such as E. coli O157:H7) [37].
Dextran-coated CdSe/ZnS quantum dots can achieve high sensitivity detection of pollutants such as phenols in water [38,39]. Quantum dots in composite silica microspheres prepared by sol-gel method are coated with cup aromatic hydrocarbon, used as luminescent probes for pesticides [40]. Single 6-mercapto-β-CD-CdTe quantum dots can be used for quantitative detection of cationic dyes like neutral red.
However, quantum dot technology still has some limitations. The preparation of quantum dots involves multiple steps (such as B-dBSA coating treatment for CdTe quantum dots, and the biotin-avidin system connection), making the process relatively complex and potentially increasing experimental costs and time. Additionally, quantum dots have certain cytotoxicity (CdTe quantum dots contain cadmium), which requires particular attention to safety when used in vivo.
3.2.3. Nano gold technology
Nanodiamond technology makes peptide detection possible by creating a metal-coated membrane that fixes the peptides to polyvinylidene fluoride [41]. The metal can bind with the functional groups of amino acids, retaining small molecules on the metal film and preventing the diffusion of peptides and small protein molecules into the metal-coated membrane during the Western Blot process. This plays a crucial role in providing new and sensitive techniques [42].
Graphene is a novel two-dimensional graphite carbon system with a single-atom layer thickness. It has garnered significant attention in the research community due to its unique structure and ability to easily bind with proteins without compromising their biological activity [43,44]. A highly sensitive WB using antibody-functionalized graphene oxide sheets and gold nanoparticles significantly reduces costs by coupling two different primary antibodies on the gold nanoparticles [45].
However, its inconvenience mainly lies in the complexity of optimizing antibody ratios. For instance, when the ratio of the auxiliary primary antibody (aAb1) to the main primary antibody (GST-Ab1) is 60/40, it can reduce costs without sacrificing signal amplification. This indicates the need for fine-tuning for different application scenarios, which increases the complexity of experimental design and operation.
Gold nanoparticles can bind to specific biomolecules (such as proteins, peptides, or nucleic acids), thereby enhancing the sensitivity and specificity of detection. In the diagnosis of avian influenza virus, gold nanoparticles are used to immobilize small peptide segments (such as the conserved peptides of NS1 protein), enabling their effective detection of in techniques like Western Blot.
Because the gold nanoparticles form a barrier to prevent peptide diffusion through the membrane, it is helpful to develop a rapid test kit with simple operation and intuitive results, which is suitable for on-site screening.
3.3. High throughput WB
Microfluidic technology involves fluid manipulation in microchannels sized from tens to hundreds of micrometers [46,47]. It is a fundamental technique for high-throughput and high-sensitivity protein detection.In figure 1, by utilizing microscale channel systems to control fluid movement, it enables sample collection, pre-treatment, injection, and separation within extremely small spaces (Figure 1(b)). A microfluidic WB method has been developed. It significantly optimizes resources and detection time [48,49]. A channel with a depth of 15 μm, width of 50 μm, and length of 8.6cm can achieve baseline separation within 8 minutes. It can resolve proteins with a 5% molecular weight difference, demonstrating high-throughput [50]. The chip outlet drags along the membrane surface. When proteins migrate from the separation channel, they directly deposit on the membrane, saving time compared to traditional WB. In Western Blotting measurements conducted under pure electronic control in individual glass microchannels, microfluidic technology has a short duration, lasting only 10-60min per run, making it suitable for multiplex analyte detection, achieving ultra-high sensitivity at the femtogram level and an extremely wide detection range [51].
Capillary Electrophoresis (CE) is an analytical separation technique capable of high-resolution separation of multiple compounds. It is particularly suitable for the separation of polar and charged compounds [52]. Systems based on capillary and Microchip Electrophoresis (MCE) have been developed to enhance the speed, automation, and mass sensitivity of Western Blotting, enabling the separation of multiple proteins in a sample. The Capillary Electrophoresis-Immunoassay (CE-IA) platform has revolutionized proteomics analysis through breakthrough microfluidic technology. Its single-well chip can integrate a nano-capillary array, allowing simultaneous separation and detection of multiple target proteins with a single injection, significantly improving throughput efficiency compared to traditional Western Blotting with 6-8 gel lanes [53].
The new technology of high-throughput WB includes heat dissipation, integrated functions and automated operation.
Because the chip format has better heat dissipation performance than the traditional capillary format, it can operate at higher electric field intensity, thus speeding up the analysis speed.
Microfabrication technologies (such as lithography and MEMS) enable the realization of complex fluid networks, allowing multiple experimental steps to be completed on a single chip, such as sample concentration, labeling, separation, and detection. When combined with electrophoresis techniques, microfluidic chips can further leverage their advantages. Additionally, by adjusting the pore size and functionality of Polyacrylamide (PA) gels through photopolymerization protocols, they can achieve different regional characteristics (such as loading zones, separation zones, and transfer zones), enabling fully integrated operations.
In addition, automated operations are implemented to reduce human interference and enhance the repeatability of experiments and the reliability of results; multiple injections from the same sample can be performed to detect different proteins, achieving multiplex detection without repeatedly stripping antibodies. From a single Jurkat cell lysate sample, 11 proteins were detected through nine injections, significantly improving detection efficiency. Each injection involves a small sample volume, enabling highly sensitive detection of trace samples, making it suitable for precious or small sample studies. Only 400ng of total protein is required for testing, compared to 10 μg with traditional methods, greatly reducing sample consumption.
High-throughput WB can be applied in proteomics analysis. Microfluidic protein blotting, by combining excellent specificity with the throughput advantage of multiplex detection, has laid the foundation for rapid proteomics localization. Studies have used microfluidic nanoliquid chromatography-tandem mass spectrometry (nanoLC-MS/MS) technology to perform proteomics analysis on airway Epithelial Lining Fluid (ELF) from patients with Chronic Obstructive Pulmonary Disease (COPD) and healthy controls. By Combining Unidirectional Polyacrylamide Gel Electrophoresis (SDS-PAGE) with intragel trypsin digestion and chip LC-MS/MS technology, approximately 300 proteins were successfully identified [54]. A team utilized microfluidic technology to transform commercial endocytic tubes into arrayed nanoreactors, achieving ultra-high quantitative analysis of up to 3000 proteins in single-cell proteomics analysis of three mammalian cell lines (HeLa, A549, U2OS) [55].
Microfluidic electrophoresis technology has demonstrated significant value in the analysis of therapeutic proteins, enabling high-precision and high-throughput detection.Studies have utilized microfluidic electrophoresis to describe the deamination products of oxytocin, achieving effective separation of all oxytocin deamination products under Specific Background Electrolyte (BGE) conditions [56].
Microfluidic technology combined with Surface Plasmon Resonance (SPR [57,58]) and standing Wave Mode (WGM) [59], among other label-free biosensing techniques, can achieve real-time, quantitative protein detection. WGM sensors have demonstrated the capability to detect viral particles (such as virions). Even when viral particles are not bound to the resonator surface, their Brownian motion can cause fluctuations in the resonance wavelength. The particle size [60] can be estimated through signal autocorrelation analysis.
Despite the high throughput and automation capabilities of microfluidic technology, its design and manufacturing processes are relatively complex. The multiplexed serpentine microchannel design requires precise alignment and optimization, increasing the difficulty of system development [61]. Additionally, the compatibility between microfluidic technology and traditional experimental methods is sometimes limited. When integrating microfluidic chips with existing Western Blotting equipment, additional adaptation or modification may be required. Microfluidic technology significantly reduces antibody usage (only 1% of that in traditional methods). However, experimental results still depend on the quality and specificity of the antibodies. If there are issues with the antibodies themselves (such as non-specific binding or cross-reactivity), it can affect the final test results [62,63]. Furthermore, signal optimization, cost, and technical barriers associated with microfluidic chip technology may limit its widespread application in laboratories or specific scenarios, requiring further improvements.
Table 1 is the comparison of characteristics and clinical value of Classic WB and novel WB.
Table 1. Comparison of characteristics and clinical value of Classic WB and novel WB
Classic WB | Spatial discrimination WB | High sensitivity WB | High-throughput WB | |
Throughput | A single experiment can only analyze a small number of samples | The throughput of a single experiment with spatially resolved WB technology is relatively low, and each microdevice can process up to about 10^3 cell. | The detection efficiency was improved, and multiple low-abundance proteins could be analyzed in a single experiment | Hundreds to thousands of samples can be analyzed simultaneously in a single experiment |
Sensitivity | The minimum detection limit of the target protein is nanogram (ng), which makes it difficult to detect ultra-low abundance proteins | The sensitivity is relatively low, limited by the spatial resolution and signal strength | It can detect proteins with picogram (pg) or even lower abundance and is suitable for rare samples or trace protein detection | High sensitivity can detect low abundance proteins, the sensitivity may be reduced due to sample complexity |
Specificity | Due to the quality of antibodies, non-specific binding is easy to occur; the specificity should be improved by optimizing conditions | Specificity depends on the quality of the antibody and spatial resolution, which may be interfered by adjacent signals | It has high specificity and can be further improved by technical improvement (such as nanoparticle enhancement) | The specificity is high, the cross-reactivity problem of multiple markers should be paid attention to |
Prime cost | The cost of equipment and reagents is low, the labor cost is high, and the overall cost is moderate | The equipment cost is medium, the reagent and consumable cost is high, and the overall cost is higher than the traditional WB | The equipment cost is moderate, special technologies and consumables (such as nanoparticles) increase the cost | The equipment cost is high, the reagent consumables are expensive, and the overall cost is significantly higher than other technologies |
Ease of operation | The operation is complicated, including sample preparation, electrophoresis, transfer | The operation is more complex, which requires accurate alignment of the microdevice and detection system to optimize the spatial resolution and signal acquisition process | The operation is relatively simple, special equipment and technical support (such as nanoblotting instrument are still required | The degree of automation is high, the experimental design and equipment debugging are complex, and the initial learning curve is steep |
Clinical value | It is mainly used for basic research and small-scale verification experiments, and has limited clinical application | Spatial resolution analysis suitable for specific samples, such as tumor tissue heterogeneity study and immune cell localization study | It is widely used in ultra-low abundance protein detection, such as cancer markers and neurotransmitter related protein research | High-throughput technology is suitable for large-scale screening, such as cancer marker detection, drug screening, liquid biopsy [64] |
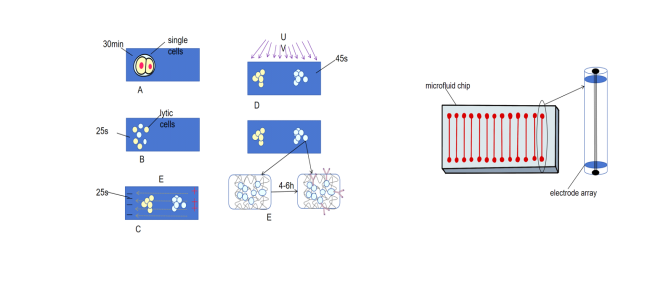
Figure 1. (a)The procedure of single-cell resolution WB. (b)the core of microfluid WB
4. Breakthrough of new qPCR technology
Due to the characteristics of low sensitivity, low throughput, non-specificity and complexity of traditional qPCR, some emerging qPCR technologies have been developed.
4.1. Ultra multiple qPCR
In figure 2, by using multi-color fluorescent probes or high-throughput design, ultra-multiplex qPCR, an emerging technology that addresses the low throughput of traditional qPCR, enables the detection of dozens to hundreds of targets in a single tube, breaking through the limitations of traditional qPCR multiplex detection (Figure 2 (a)). The innovation of ultra multiplex qPCR mainly lies in TaqMan probes and molecular beacon technology.
Multiplex real-time fluorescent PCR technology based on TaqMan probes has low cost (each test costs only $1.3) and is rapidly simple. For example, it can complete species identification within 2 hours in a single reaction tube, with 50 minutes for nucleic acid extraction and 70 minutes for nucleic acid amplification. Multiplex PCR is also an effective tool for identifying major mycobacterial pathogens and has shown good diagnostic performance in clinical samples. However, multiplex qPCR still has certain limitations; for some samples, it still has limitations in sensitivity and specificity, which restricts the development of multiplex qPCR. Currently, there are fewer sample types suitable for multiplex qPCR measurements [65].
Molecular beacons are also a crucial technology for achieving ultra-multiplex qPCR. The tail of the molecular beacon contains a fluorophore and a quencher group, normally remaining free in the system. When the two groups are close together, the quencher group quenches the fluorescence of the fluorophore, preventing the blank solution from emitting light. The intensity of the fluorescence indicates the amount of amplified DNA. Some studies have used unique combinations of two fluorescent colors to label molecular beacon probes for multiplex PCR screening and analysis, increasing the number of detectable targets while maintaining high specificity and sensitivity [66].
In tumor diagnosis and treatment, ultra-multiplex qPCR can be used for multi-gene mutation screening, rapid detection of tumor-related gene mutations, and provide a basis for targeted therapy. For example, in the detection of lung cancer patients, multiple gene mutations of EGFR can be detected simultaneously, helping doctors select appropriate targeted drugs [67].
Immunome analysis requires the detection of a large number of immune cell genes, and ultra-multiplex qPCR can efficiently complete this task, aiding immunological research and immunotherapy for diseases. Studies have shown that high-throughput sequencing technology in immunome research primarily focuses on infectious diseases, it also emphasizes the efficiency of high-throughput sequencing in analyzing immune cell genes [68]. This provides a technical development background and relevant ideas for the application of ultra-multiplex qPCR in immunome analysis, offering valuable insights into technological trends.
Ultra multiplex qPCR can simultaneously detect multiple microbial genes, making it useful for monitoring environmental microbial pathogens and ecological research. By designing specific primers and probes targeting different microbial groups, this method achieves quantitative analysis of multiple target microbial pathogens in a single reaction system. A study introduced a multiplex real-time qPCR method for detecting and quantifying Pythium tracheiphilum in soil, which not only detects the presence of pathogens but also quantifies them, aiding in better risk assessment [69].
However, multiplex qPCR is prone to reagent competition issues. In multiple reactions, different amplification products compete for reaction reagents (such as DNA polymerase, dNTPs, buffers, and MgCl2), which can lead to reduced amplification efficiency of certain target sequences. If the concentration of the target DNA is unknown or varies significantly, high-abundance targets may consume more reagents, thereby inhibiting the amplification [70] of low-abundance targets. Additionally, primer and probe design for multiplex qPCR requires consideration of specificity for multiple targets and annealing temperatures, making it more challenging to design.
4.2. Digital qPCR
Digital qPCR (dPCR) is a novel quantitative nucleic acid (DNA, cDNA, or RNA) technology. It uses limited dilution and the Poisson distribution to analyze data and determine the absolute copy number concentration of the target.In Figure 2, a complete qPCR reaction system is divided into several (typically tens of thousands to millions of) independent reaction zones (nanoliter volumes). After PCR amplification, the reaction zones containing the target DNA emit a fluorescent signal, indicating a positive result; those without the target DNA show a negative result indicate a positive result. Finally, the copy number concentration of the target DNA is calculated based on the proportion of negative reaction zones using the Poisson distribution (Figure 2(b)). Unlike real-time quantitative PCR, the entire digital PCR process does not require an amplification standard curve or housekeeping genes, offering excellent accuracy and reproducibility, enabling true absolute quantification [71].
dPCR divides the reaction system into tens of thousands of droplets, each amplifying independently. The absolute copy number is calculated based on the proportion of positive droplets, eliminating the need for a standard curve to achieve absolute quantification. Research has focused on the potential applications of droplet-based digital PCR (ddPCR) and optimized next-generation sequencing (NGS) in detecting ctDNA, for cancer recurrence, minimal residual disease detection, and early diagnosis of cancer patients. Compared to tumor tissue biopsies, blood-based ctDNA analysis is minimally invasive and facilitates regular follow-up for cancer patients, better reflecting the patient's pathological condition and helping to highlight tumor heterogeneity and multiple tumor sites [72].
Droplet-based and chip-based technologies are the two major techniques in digital qPCR. Droplet-based ddPCR is based on oil-in-water droplet generation technology [73]. Chip-baseddPCR utilizes microfluidic chips to segment reaction units, each with its own advantages. Microfluidic chips, which are used in dPCR and other technologies, excel in detecting pathogens and gene mutations, as well as in disease prevention and diagnosis, demonstrating their advantages in precise control of reaction conditions in nucleic acid testing [74]. On the other hand, ddPCR is suitable for high-throughput detection, allowing researchers to choose the appropriate platform according to experimental needs.
In virology research and clinical diagnosis, dPCR can be used for precise detection of viral loads, such as HIV and HPV, providing a basis for disease diagnosis, treatment, and prognosis assessment. During the treatment of HIV patients, dPCR is used to accurately monitor changes in viral load, evaluate treatment effectiveness, and adjust treatment plans. A study compared the accuracy of total HIV DNA quantification using real-time PCR (qPCR) and digital PCR (dPCR) in patients who have been on antiretroviral therapy for a long time. The study found that both methods showed high correlation, dPCR had better repeatability and reproducibility, allowing for more accurate monitoring of HIV reservoir [75].
Although the data analysis of dPCR is relatively simple, the complexity of data analysis is still high when dealing with complex samples and multiple detection, which requires special instruments and equipment, high cost, cumbersome operation and long time, etc., which limits its application.
4.3. Microfluidic qPCR
Microfluidic qPCR and ultra-multiplex qPCR both offer high throughput, but they operate on different principles. In figure 2, microfluidic qPCR integrates microfluidic chip technology to downsize and automate reaction systems, thereby boosting detection efficiency and accuracy (Figure 2(c)). These integrated chips consolidate multiple reaction chambers into a single unit, facilitating high-throughput testing, as exemplified by the GeneXpert platform [76]. Portable devices, on the other hand, are tailored for rapid on-site testing, such as the POCT [77] for pathogens. Integrated chips are particularly advantageous for large-scale sample testing, while portable devices are indispensable for emergency testing and on-site diagnosis, catering to diverse testing requirements.
Once the sample is loaded into the wells, the chip undergoes heating and cooling at precise thermal cycling temperatures. The CF-PCR chip transports the sample to a fixed temperature zone to achieve thermal cycling. CF-PCR chips can be further classified into various design types, including serpentine channel devices, spiral microchannel devices, oscillating devices, closed-loop devices, and straight channel devices. These design variations enable microfluidic PCR devices to be extensively used in molecular biology applications like gene detection and molecular sequencing.
In personalized medical settings, microfluidic qPCR excels, particularly in bedside genetic testing. It provides patients with swift and precise diagnostic results, supporting targeted treatment. For cancer patients, microfluidic qPCR can promptly detect tumor marker genes at the bedside, offering timely data for adjusting treatment plans. For instance, a study developed a digital PCR chip based on microfluidic technology that detected lung cancer-specific tumor marker lncRNA in exosomes from saliva. This method boasts a low detection limit of 10 copies/μL and high sensitivity, paving the way for innovative tumor marker detection methods.
However, microfluidic qPCR demands meticulous sample processing, necessitating rigorous nucleic acid extraction and purification. Inadequate sample handling can compromise test accuracy. For example, Fluidigm testing has shown heightened sensitivity in detecting serotype 5 and serogroup 25AF/38, which may result in false-positives [78]. Moreover, the cost of microfluidic qPCR equipment and reagents is relatively high, restricting its widespread adoption in resource-constrained areas. The operational complexity also requires specialized technical personnel for management and maintenance.
Table 2 is the comparison of several common qPCR techniques.
Table 2. Comparison of several common qPCR techniques
Classic qPCR | Super-multiplet qPCR | Digital qPCR | Microfluidic qPCR | |||
Throughput | Low throughput | Through multi-color fluorescent probes or high-throughput design, tens to hundreds of targets can be detected in a single tube, breaking through the limitations of traditional multiple detection | - | Combined with microfluidic chip technology, the reaction system is miniaturized and automated to support high-throughput detection | ||
Sensitivity | - | The molecular beacon increases the number of detectable targets and has high specificity and sensitivity | It has good accuracy and reproducibility, and high detection sensitivity | - | ||
Specificity | - | Some samples have low specificity | - | - | ||
Ease of operation | relatively simple | The design of primers and probes requires consideration of multiple objectives, which is difficult to design | Data analysis is complex and cumbersome when dealing with complex samples and multiple tests. | The sample processing requirements are high, and professional technical personnel are required to operate and maintain | ||
Prime cost | Relatively low | The technology cost of TaqMan probes is low (US $1.3 per reagent) | The equipment and operation costs are high | Equipment and reagents are expensive | ||
Clinical application | Gene quantitative analysis and other routine tests | Screening for multi-gene mutations in tumors, immunogenetic analysis, and detection of environmental microbial communities | Precise detection of viral load (HIV), determination of copy number of target gene of transgenic organisms [79], cancer-related detection | Personalized treatment bedside genetic testing, tumor marker gene rapid detection | ||
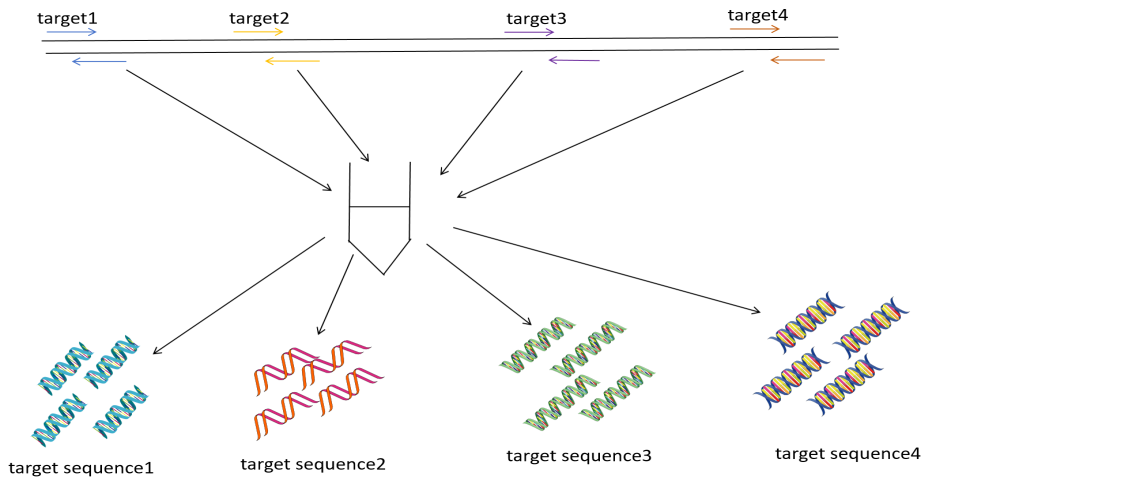
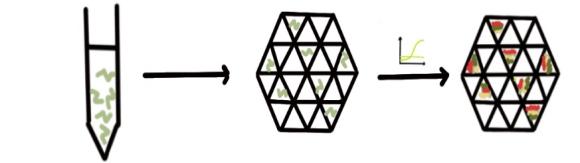
Figure 2. (a) the procedure of ultra-multiplex qPCR (b) the procedure of dPCR (c) the procedure of microfluid qPCR
4.4. Other emerging qPCR technologies
Other burgeoning qPCR technologies, such as CRISPR-qPCR, single molecule qPCR, and LAMP-qPCR, are broadening the horizons of qPCR applications. The CRISPR-Cas9 system, a potent genomic editing instrument widely employed in gene function research, disease treatment, and biotechnology advancement [80], underpins CRISPR-qPCR technology. By harnessing the heightened specificity of the CRISPR-Cas system, CRISPR-qPCR can detect low-abundance nucleic acids, thereby extending the reach of qPCR applications [81]. In the context of viral variant detection, CRISPR-qPCR excels at pinpointing specific mutation sites, enhancing both the specificity and sensitivity of the detection process.
Single-molecule qPCR, on the other hand, can detect individual molecule templates, surmounting the sensitivity constraints of traditional qPCR and offering a novel approach for the detection of ultra-low abundance nucleic acids. For instance, when analyzing gene expression in scarce cell types, single-molecule qPCR can precisely quantify gene expression levels within single cells, shedding light on cellular heterogeneity [82].
Isothermal amplification techniques, exemplified by LAMP-qPCR, streamline the operational process by reducing thermal cycling steps. This makes LAMP-qPCR particularly well-suited for resource-constrained environments and rapid on-site testing. In remote areas or primary healthcare settings, LAMP-qPCR can swiftly detect pathogens under simple conditions, thereby improving access to testing [83]. These innovative qPCR technologies are collectively pushing the boundaries of what is achievable, unlocking new possibilities for disease detection, diagnosis, and research.
5. Conclution
With the advent of novel WB and qPCR technologies, they have attracted growing attention due to their distinctive methodologies and swift, convenient detection capabilities, potentially unlocking fresh avenues for disease detection, diagnosis, treatment, and efficacy monitoring. The triangular validation approach integrating WB, qPCR, and mass spectrometry facilitates the assessment of both protein and gene expression levels, furnishing multi-faceted data support for research endeavors and markedly bolstering the reliability of experimental outcomes. Researchers have harnessed LC-MS/MS to pinpoint proteins interacting with LKB1 in lung cancer and corroborated the influence of LKB1 deficiency on lung cancer progression through qPCR and WB techniques, thereby rendering the findings more compelling [84]. Single-cell multi-omics analysis ingeniously fuses traditional comet assays (for DNA analysis) with immunoblotting (for protein analysis), enabling the concurrent acquisition of DNA and protein information from individual cells [85]. SplitBlot leverages microfluidic precision separation to isolate proteins and genomic DNA, encapsulating DNA within agarose microwells for protein scrutiny, extracting DNA for PCR, and capturing proteins onto a gel for detection. This approach achieves both DNA amplification and protein separation, offering tools to correlate genotype and protein expression at the single-cell level [86]. Microfluidic technology excels in efficiently separating and enriching target biomarkers (such as circulating tumor DNA, circulating tumor cells, and exosomes) from intricate biological samples via precise fluid management and separation techniques. This is particularly advantageous for early cancer screening and the dynamic monitoring of disease progression during treatment. By combining ddPCR with RNA sequencing, researchers can identify several significantly differentially expressed GPCR genes, serving as potential drug targets for a rat model of cardiac dysfunction induced by therapeutic stress overload [87]. Harnessing these strengths, the new technology promises to deliver low-cost and widely accessible solutions for early clinical testing and drug development.
Nevertheless, these emerging detection technologies also encounter certain hurdles. For example, in the detection of biomarkers using microfluidic chips, the size range and physicochemical properties of biomarkers like CTCs, ctDNA, and exosomes exhibit substantial variability. A one-size-fits-all approach is infeasible; tailored microfluidic techniques must be devised for each biomarker. Currently, this system remains in the laboratory phase, lacking large-scale clinical trials to validate its practicality and enhance patient outcomes. Collaboration with medical professionals is crucial to provide pragmatic insights for clinical deployment. Consequently, it is imperative to synergize physical methods (including inertial, magnetic, acoustic, and optical techniques) with biochemical approaches to isolate and analyze biomarkers, thereby augmenting clinical trials. Spatially resolved WB technology also grapples with limitations in throughput and sensitivity. Thus, concerted efforts are needed from multiple fronts, encompassing throughput enhancement, detection limit reduction, optimization of gel-based immuno-probes, adjustment of PAGE conditions, achievement of multiplexed detection, and refinement of micro-device fabrication.
In conclusion, despite the challenges presented by novel WB and qPCR technologies, technological progress and interdisciplinary cooperation can significantly propel their development. The application of these cutting-edge technologies will amplify research efficiency, influencing both fundamental research and clinical applications, pushing the frontiers of scientific exploration, and steering us towards more precise healthcare.
References
[1]. Moritz, C. P. (2020). 40 years Western Blotting: A Scientific Birthday Toast. Journal of Proteomics, 212, 103575. https://doi.org/10.1016/j.jprot.2019.103571
[2]. Begum, H., Murugesan, P., & Tangutur, A. D. (2022). Western Blotting: a Powerful Staple in Scientific and Biomedical Research. Biotechniques, 73(1), 58–69. https://doi.org/10.2144/btn-2022-0003
[3]. Singh, K. K., Gupta, A., Bharti, C., & Sharma, H. (2021). Emerging Techniques of Western Blotting for Purification and Analysis of Protein. Future Journal of Pharmaceutical Sciences, 7(1). https://doi.org/10.1186/s43094-021-00386-1
[4]. Yang, P., & Mahmood, T. (2012). Western blot: Technique, Theory, and Trouble Shooting. North American Journal of Medical Sciences, 4(9), 429. https://doi.org/10.4103/1947-2714.100998
[5]. Towbin, H., Staehelin, T., & Gordon, J. (1979). Electrophoretic Transfer of Proteins from Polyacrylamide Gels to Nitrocellulose Sheets: Procedure and Some Applications. Proceedings of the National Academy of Sciences of the United States of America, 76(9), 4350–4354. https://doi.org/10.1073/pnas.76.9.4350
[6]. Litovchick, L. (2018). Immunoblotting: Transfer of Proteins from Gels to Membranes. Cold Spring Harbor Protocols, 2018(10), pdb.prot98442. https://doi.org/10.1101/pdb.prot098442
[7]. Sule, R., Rivera, G., & Gomes, A. V. (2023). Western Blotting (Immunoblotting): History, Theory, Uses, Protocol and Problems. Biotechniques, 75(3), 99–114. https://doi.org/10.2144/btn-2022-0034
[8]. Heidebrecht, F., Heidebrecht, A., Schulz, I., Behrens, S. E., & Bader, A. (2009). Improved Semiquantitative Western Blot Technique with Increased Quantification Range. Journal of Immunological Methods, 345(1–2), 40–48. https://doi.org/10.1016/j.jim.2009.02.005
[9]. Bose, U., Wijffels, G., Howitt, C. A., & Colgrave, M. L. (2019). Proteomics: Tools of the Trade. In J. L. Capelo-Martínez (Ed.), Emerging Sample Treatments in Proteomics (pp. 1–22). Springer. https://doi.org/10.1007/978-3-030-12298-0_1
[10]. Han, X. L., Yang, J. R., Bao, F. K., & Liu, A. H. (2017). Progress of Stain-Free Technology and Total Protein Analysis in Western Blotting. Letters in Biotechnology, 28(5), 709–714. https://doi.org/10.3969/j.issn.1009-0002.2017.05.028
[11]. Lallier, S. W., Hill, C. L., Nichols, D. P., & Reynolds, S. D. (2019). Protein Abundance Determination: An Optimized Western Blot Workflow. Annals of Clinical and Laboratory Science, 49(4), 507–512.
[12]. Kong, W., Li, Y., Cheng, S., Yan, C., An, S., Dong, Z., Yan, L., & Yuan, Y. (2016). Luminex xMAP Combined with Western Blot Improves HIV Diagnostic Sensitivity. Journal of Virological Methods, 227, 1–5. https://doi.org/10.1016/j.jviromet.2015.10.019
[13]. Mullis, K., Faloona, F., Scharf, S., Saiki, R., Horn, G., & Erlich, H. (1986). Specific Enzymatic Amplification of DNA in Vitro: the Polymerase Chain Reaction. Cold Spring Harbor Symposia on Quantitative Biology, 51(Pt 1), 263–273. https://doi.org/10.1101/sqb.1986.051.01.032
[14]. Smith, C. J., & Osborn, A. M. (2009). Advantages and Limitations of Quantitative PCR (Q-PCR)-based Approaches in Microbial Ecology. FEMS Microbiology Ecology, 67(1), 6–20. https://doi.org/10.1111/j.1574-6941.2008.00629.x
[15]. Tajadini, M., Panjehpour, M., & Javanmard, S. (2014). Comparison of SYBR Green and TaqMan Methods in Quantitative Real-time Polymerase Chain Reaction Analysis of Four Adenosine Receptor Subtypes. Advanced Biomedical Research, 3(1), 85. https://doi.org/10.4103/2277-9175.127998
[16]. Li, B., & Yan, T. (2021). Next Generation Sequencing Reveals Limitation of QPCR Methods in Quantifying Emerging Antibiotic Resistance Genes (ARGs) in the Environment. Applied Microbiology and Biotechnology, 105(7), 2925–2936. https://doi.org/10.1007/s00253-021-11202-6
[17]. Zhao, K., Shi, K., Zhou, Q., Xiong, C., Mo, S., Zhou, H., Long, F., Wei, H., Hu, L., & Mo, M. (2022). The Development of a Multiplex Real-Time Quantitative PCR Assay for the Differential Detection of the Wild-Type Strain and the MGF505-2R, EP402R and I177L Gene-Deleted Strain of the African Swine Fever Virus. Animals, 12(14), 1754. https://doi.org/10.3390/ani12141754
[18]. Zhang, Q., Wang, J., Deng, F., Yan, Z., Xia, Y., Wang, Z., Ye, J., Deng, Y., Zhang, Z., Qiao, M., Li, R., Denduluri, S. K., Wei, Q., Zhao, L., Lu, S., Wang, X., Tang, S., Liu, H., Luu, H. H., Haydon, R. C., He, T. C., & Jiang, L. (2015). TqPCR: A Touchdown qPCR Assay with Significantly Improved Detection Sensitivity and Amplification Efficiency of SYBR Green qPCR. PLOS ONE, 10(7), e0132666. https://doi.org/10.1371/journal.pone.0132666
[19]. Hu, X., Zang, X., & Lv, Y. (2021). Detection of Circulating Tumor Cells: Advances and Critical Concerns. Oncology Letters, 21(5), 422. https://doi.org/10.3892/ol.2021.12992
[20]. Hermann-Bank, M. L., Skovgaard, K., Stockmarr, A., Larsen, N., & Molbak, L. (2013). The Gut Microbiotassay: a High-Throughput QPCR Approach Combinable with Next Generation Sequencing to Study Gut Microbial Diversity. BMC Genomics, 14, 788. https://doi.org/10.1186/1471-2164-14-788
[21]. Logan, J. M. J., & Edwards, K. J. (2004). An Overview of Real-Time PCR Platforms. https://doi.org/10.1007/978-1-4020-2400-9_2
[22]. Kang, C., Yamauchi, K. A., Vlassakis, J., Sinkala, E., Duncombe, T. A., & Herr, A. E. (2016). Single Cell–Resolution Western Blotting. Nature Protocols, 11(8), 1508–1530. https://doi.org/10.1038/nprot.2016.089
[23]. Hughes, A. J., Spelke, D. P., Xu, Z., Kang, C., Schaffer, D. V., & Herr, A. E. (2014). Single-cell Western Blotting. Nature Methods, 11(7), 749–755. https://doi.org/10.1038/nmeth.2992
[24]. Kang, C., Lin, J. G., Xu, Z., Kumar, S., & Herr, A. E. (2014). Single-Cell Western Blotting after Whole-Cell Imaging to Assess Cancer Chemotherapeutic Response. Analytical Chemistry, 86(20), 10429–10436. https://doi.org/10.1021/ac502932t
[25]. Yamauchi, K. A., & Herr, A. E. (2017). Subcellular Western Blotting of Single Cells. Microsystems & Nanoengineering, 3(1), 1–10. https://doi.org/10.1038/micronano.2016.79
[26]. Li, J. J., Bickel, P. J., & Biggin, M. D. (2014). System Wide Analyses Have Underestimated Protein Abundances and the Importance of Transcription in Mammals. PeerJ, 2, e270. https://doi.org/10.7717/peerj.270
[27]. Durmus, N. G., Tekin, H. C., Guven, S., Sridhar, K., Arslan Yildiz, A., Calibasi, G., Ghiran, I., Davis, R. W., Steinmetz, L. M., & Demirci, U. (2015). Magnetic Levitation of Single Cells. Proceedings of the National Academy of Sciences of the United States of America, 112(28), 8606–8611. https://doi.org/10.1073/pnas.1509250112
[28]. Karin, M., & Greten, F. R. (2005). NF-κB: Linking Inflammation and Immunity to Cancer Development and Progression. Nature Reviews Immunology, 5(10), 749–759. https://doi.org/10.1038/nri1703
[29]. Zeder-Lutz, G., Cherouati, N., Reinhart, C., Pattus, F., & Wagner, R. (2006). Dot-Blot Immunodetection as a Versatile and High-Throughput Assay to Evaluate Recombinant GPCRs Produced in the Yeast Pichia Pastoris. Protein Expression and Purification, 50(1), 118–127. https://doi.org/10.1016/j.pep.2006.03.013
[30]. Ortega Ibarra, J. M., Cifuentes-Castro, V. H., Medina-Ceja, L., & Morales-Villagrán, A. (2021). Nano Dot Blot: An Alternative Technique for Protein Identification and Quantification in a High Throughput Format. Journal of Neuroscience Methods, 358, 109194. https://doi.org/10.1016/j.jneumeth.2021.109194
[31]. Klabunde, T., & Hessler, G. (2002). Drug Design Strategies for Targeting G-protein-coupled Receptors. ChemBioChem, 3(10), 928–944. https://doi.org/10.1002/1439-7633(20021004)3:10<928::AID-CBIC928>3.0.CO;2-5
[32]. Scholl, B., Liu, H. Y., Long, B. R., McCarty, O. J., O'Hare, T., Druker, B. J., & Vu, T. Q. (2009). Single Particle Quantum Dot Imaging Achieves Ultrasensitive Detection Capabilities for Western Immunoblot Analysis. ACS Nano, 3(6), 1318–1328. https://doi.org/10.1021/nn900390t
[33]. Bakalova, R., Zhelev, Z., Ohba, H., & Baba, Y. (2005). Quantum Dot-Based Western Blot Technology for Ultrasensitive Detection of Tracer Proteins. Journal of the American Chemical Society, 127(26), 9328–9329. https://doi.org/10.1021/ja0510055
[34]. Wald, F. V. (1977). Applications of CdTe. A Review. Revue de Physique Appliquée, 12(2), 277–290. https://doi.org/10.1051/rphysap:01977001202027700
[35]. Chen, W., Xu, D., Liu, L., Peng, C., Zhu, Y., Ma, W., Bian, A., Li, Z., Jin, Z., Zhu, S. (2009). Ultrasensitive Detection of Trace Protein by Western Blot Based on POLY-Quantum Dot Probes. Analytical Chemistry, 81(21), 9194–9198. https://doi.org/10.1021/ac901429a
[36]. Allen, S. J., & Dawbarn, D. (2006). Clinical Relevance of the Neurotrophins and Their Receptors. Clinical Science, 110(2), 175–191. https://doi.org/10.1042/CS20050347
[37]. Su, X. L., & Li, Y. (2004). Quantum Dot Biolabeling Coupled with Immunomagnetic Separation for Detection of Escherichia Coli O157:H7. Analytical Chemistry, 76(16), 4806–4810. https://doi.org/10.1021/ac049513k
[38]. Li, H., & Han, C. (2008). Sonochemical Synthesis of Cyclodextrin-Coated Quantum Dots for Optical Detection of Pollutant Phenols in Water. Chemistry of Materials, 20(19), 6053–6059. https://doi.org/10.1021/cm8009176
[39]. Wang, L., Liu, S., Peng, J., & He, Y. (2010). The Synthesis of Mono-6-Thio-β-Cyclodextrin Capped CdTe QDs and Its Interaction with Neutral Red. Science China Chemistry, 53(6), 1358–1365. https://doi.org/10.1007/s11426-010-3092-2
[40]. Li, H., & Qu, F. (2007). Synthesis of CdTe Quantum Dots in Sol−Gel-Derived Composite Silica Spheres Coated with Calix[4]arene as Luminescent Probes for Pesticides. Chemistry of Materials, 19(17), 4148–4154. https://doi.org/10.1021/cm0700089
[41]. Duchesne, L., & Fernig, D. G. (2007). Silver and Gold Nanoparticle-Coated Membranes for Femtomole Detection of Small Proteins and Peptides by Dot and Western Blot. Analytical Biochemistry, 362(2), 287–289. https://doi.org/10.1016/j.ab.2006.09.014
[42]. Emami, T., Madani, R., Rezayat, S. M., Golchinfar, F., & Sarkar, S. (2012). Applying of Gold Nanoparticle to Avoid Diffusion of the Conserved Peptide of Avian Influenza Nonstructural Protein from Membrane in Western Blot. Journal of Applied Poultry Research, 21(3), 563–566. https://doi.org/10.3382/japr.2011-00343
[43]. Shen, J., Hu, Y., Shi, M., Li, N., Ma, H., & Ye, M. (2010). One Step Synthesis of Graphene Oxide−Magnetic Nanoparticle Composite. The Journal of Physical Chemistry C, 114(3), 1498–1503. https://doi.org/10.1021/jp909756r
[44]. Green, A. A., & Hersam, M. C. (2010). Emerging Methods for Producing Monodisperse Graphene Dispersions. The Journal of Physical Chemistry Letters, 1(2), 544–549. https://doi.org/10.1021/jz900235f
[45]. Lin, H., Huo, J., Zhang, A., Liu, Y., Wang, Q., Cai, Y., Ying, W., Qin, W., Zhang, Y., & Qian, X. (2012). A Sensitive Dual Signal Amplification Method for Western Blotting Based On Antibody-Functionalised Graphene Oxide and Gold Nanoparticles. Analyst, 137(16), 3620–3623. https://doi.org/10.1039/C2AN36200A
[46]. Wu, D., Qin, J., & Lin, B. (2008). Electrophoretic Separations on Microfluidic Chips. Journal of Chromatography A, 1184(1–2), 542–559. https://doi.org/10.1016/j.chroma.2007.12.016
[47]. Whitesides, G. M. (2006). The Origins and The Future of Microfluidics. Nature, 442(7101), 368–373. https://doi.org/10.1038/nature05058
[48]. Kurien, B. T., & Scofield, R. H. (Eds.). (2015). Western Blotting: Methods and Protocols (Methods in Molecular Biology, Vol. 1312). Humana Press.
[49]. Sanders, B. J., Kim, D. C., & Dunn, R. C. (2016). Recent Advances in Microscale Western Blotting. Analytical Methods, 8(39), 7002–7013. https://doi.org/10.1039/C6AY01947A
[50]. Jin, S., Furtaw, M. D., Chen, H., Lamb, D. T., Ferguson, S. A., Arvin, N. E., Dawod, M., & Kennedy, R. T. (2016). Multiplexed Western Blotting Using Microchip Electrophoresis. Analytical Chemistry, 88(13), 6703–6710. https://doi.org/10.1021/acs.analchem.6b00705
[51]. Hughes, A. J., & Herr, A. E. (2012). Microfluidic Western blotting. Proceedings of the National Academy of Sciences of the United States of America, 109(52), 21450–21455. https://doi.org/10.1073/pnas.1214643109
[52]. Ramautar, R., Demirci, A., & Jong, G. J. D. (2006). Capillary Electrophoresis in Metabolomics. TrAC Trends in Analytical Chemistry, 25(5), 455–466. https://doi.org/10.1016/j.trac.2006.03.003
[53]. Mishra, M., Tiwari, S., & Gomes, A. V. (2017). Protein Purification and Analysis: Next Generation Western Blotting Techniques. Expert Review of Proteomics, 14(11), 1037–1053. https://doi.org/10.1080/14789450.2017.1388167
[54]. Franciosi, L., Govorukhina, N., Fusetti, F., Poolman, B., Lodewijk, M. E., Timens, W., Postma, D., ten Hacken, N., & Bischoff, R. (2013). Proteomic Analysis of Human Epithelial Lining Fluid by Microfluidics‐Based NanoLC‐MS/MS: A Feasibility Study. Electrophoresis, 34(18), 2683–2694. https://doi.org/10.1002/elps.201300020
[55]. Wang, Y., Guan, Z. Y., Shi, S. W., Jiang, Y. R., Zhang, J., Yang, Y., Wu, Q., Wu, J., Chen, J. B., Ying, W. X., Xu, Q. Q., Fan, Q. X., Wang, H. F., Zhou, L., Wang, L., Fang, J., Pan, J. Z., & Fang, Q. (2024). Pick-up Single-Cell Proteomic Analysis for Quantifying up to 3000 Proteins in a Mammalian Cell. Nature Communications, 15(1), 1–13. https://doi.org/10.1038/s41467-024-45659-4
[56]. Creamer, J. S., Oborny, N. J., & Lunte, S. M. (n.d.). Recent Advances in the Analysis of Therapeutic Proteins by Capillary and Microchip Electrophoresis. Analytical Methods. https://doi.org/10.1039/C7AY00841B
[57]. Vollmer, F., & Arnold, S. (2008). Whispering-Gallery-Mode Biosensing: Label-Free Detection Down to Single Molecules. Nature Methods, 5(7), 591–596. https://doi.org/10.1038/nmeth.1221
[58]. Hunt, H. K., & Armani, A. M. (2010). Label-Free Biological and Chemical Sensors. Nanoscale, 2(9), 1544. https://doi.org/10.1039/C0NR00201A
[59]. Wang, Z., Zhou, B., & Zhang, A. P. (2024). High-Q WGM Microcavity-Based Optofluidic Sensor Technologies for Biological Analysis. Biomicrofluidics, 18(4), 041502. https://doi.org/10.1063/5.0175538
[60]. Arnold, S., Ramjit, R., Keng, D., Kolchenko, V., & Teraoka, I. (2008). Micro Particle Photophysics Illuminates Viral Bio-sensing. Faraday Discussions, 137, 65–83. https://doi.org/10.1039/B702920A
[61]. He, S., Zhang, Y., Wang, P., Xu, X., Zhu, K., Pan, W., Liu, W., Cai, K., Sun, J., Zhang, W., & Jiang, X. (2015). Multiplexed Microfluidic Blotting of Proteins and Nucleic Acids by Parallel, Serpentine Microchannels. Lab on a Chip, 15(1), 105–112. https://doi.org/10.1039/C4LC01167J
[62]. Jensen, B. C., Swigart, P. M., & Simpson, P. C. (2009). Ten Commercial Antibodies for Alpha-1-Adrenergic Receptor Subtypes are Nonspecific. Naunyn-Schmiedeberg's Archives of Pharmacology, 379(4), 409–412. https://doi.org/10.1007/s00210-008-0368-6
[63]. Michel, M. C., Wieland, T., & Tsujimoto, G. (2009). How Reliable are G-Protein-Coupled Receptor Antibodies? Naunyn-Schmiedeberg's Archives of Pharmacology, 379(4), 385–388. https://doi.org/10.1007/s00210-009-0395-y
[64]. Surappa, S., Multani, P., Parlatan, U., Sinawang, P. D., Kaifi, J., Akin, D., & Demirci, U. (2024). Integrated “Lab-on-a-Chip” Microfluidic Systems for Isolation, Enrichment, and Analysis of Cancer Biomarkers. Lab Chip, 23(13), 2942–2958. https://doi.org/10.1039/D4LC00314K
[65]. Fang, T., Peng, L., Yang, T., Cai, Q., Li, H., Li, H., & Cai, L. (2025). Development and Evaluation of Multiplex Real-Time PCR for Rapid Identifying Major Pathogenic Mycobacteria from Pulmonary and Extrapulmonary Clinical Samples in Eastern China. Heliyon, 11(1), e41384. https://doi.org/10.1016/j.heliyon.2024.e41384
[66]. Marras, S. A. E., Tyagi, S., Antson, D., & Kramer, F. R. (2019). Color-Coded Molecular Beacons for Multiplex PCR Screening Assays. PLoS One, 14(3), e213906. https://doi.org/10.1371/journal.pone.0213906
[67]. Lin, C., Yeh, K., Chang, Y., Hsu, N. C., & Chang, J. (2010). Rapid Detection of Epidermal Growth Factor Receptor Mutations with Multiplex PCR and Primer Extension in Lung Cancer. Journal of Biomedical Science, 17(1), 37. https://doi.org/10.1186/1423-0127-17-37
[68]. Hou, D., Chen, C., Seely, E. J., Chen, S., & Song, Y. (2016). High-Throughput Sequencing-Based Immune Repertoire Study during Infectious Disease. Frontiers in Immunology, 7, 336. https://doi.org/10.3389/fimmu.2016.00336
[69]. Van der Heyden, H., Wallon, T., Lévesque, C. A., & Carisse, O. (2019). Detection and Quantification of Pythium tracheiphilum in Soil by Multiplex Real-Time qPCR. Plant Disease, 103(3), 475–483. https://doi.org/10.1094/PDIS-03-18-0419-RE
[70]. Eckford-Soper, L. K., & Daugbjerg, N. (2015). Development of a Multiplex Real-Time qPCR Assay for Simultaneous Enumeration of up to Four Marine Toxic Bloom-Forming Microalgal Species. Harmful Algae, 48, 37–43. https://doi.org/10.1016/j.hal.2015.05.006
[71]. Lin, J., Su, G., Su, W., & Zhou, C. (2017). Progress in digital PCR technology and application. Chinese Journal of Biotechnology, 33(2), 170–177. https://doi.org/10.13345/j.cjb.160269
[72]. Postel, M., Roosen, A., Laurent-Puig, P., Taly, V., & Wang-Renault, S. (2018). Droplet-Based Digital PCR and Next Generation Sequencing for Monitoring Circulating Tumor DNA: A Cancer Diagnostic Perspective. Expert Review of Molecular Diagnostics, 18(1), 7–17. https://doi.org/10.1080/14737159.2017.1394582
[73]. Pawinwongchai, J. (2020). Study of Platelet Production from Megakaryocyte by Using Induced Pluripotent Stem Cell. Chulalongkorn University Theses and Dissertations, 442. https://digital.car.chula.ac.th/chulaetd/442
[74]. Qu, Y. L. (2019). Research on the Detection of Salivary Exosome Tumor Markers by Digital PCR Chip [Master’s thesis]. Dalian Medical University.
[75]. Renault, C., Bolloré, K., Pisoni, A., Motto-Ros, C., Van de Perre, P., Reynes, J., & Tuaillon, E. (2022). Accuracy of Real-Time PCR and Digital PCR for the Monitoring of Total HIV DNA under Prolonged Antiretroviral Therapy. Scientific Reports, 12(1), 1–10. https://doi.org/10.1038/s41598-022-13581-8
[76]. Banada, P. P., Deshpande, S., Banik, S., Shah, D., Koshy, R., Patel, B., Kwiatkowski, R., Persing, D., & Alland, D. (2019). Multiplex Detection of Three Select Agents Directly from Blood by Use of the GeneXpert System. Journal of Clinical Microbiology, 57(5), e00036-19. https://doi.org/10.1128/JCM.00036-19
[77]. Bai, H., Liu, Y., Gao, L., Wang, T., Zhang, X., Hu, J., Ding, L., Zhang, Y., Wang, Q., Wang, L., Li, J., Zhang, Z., Wang, Y., Shen, C., Ying, B., Niu, X., & Hu, W. (2024). A portable All-In-One Microfluidic Device with Real-Time Colorimetric LAMP for HPV16 and HPV18 DNA Point-of-Care Testing. Biosensors and Bioelectronics, 248, 115968. https://doi.org/10.1016/j.bios.2023.115968
[78]. Olwagen, C. P., Adrian, P. V., & Madhi, S. A. (2019). Performance of the Biomark HD Real-Time qPCR System (Fluidigm) for the Detection of Nasopharyngeal Bacterial Pathogens and Streptococcus Pneumoniae Typing. Scientific Reports, 9(1), 1–10. https://doi.org/10.1038/s41598-019-42846-y
[79]. You, Y. H., Zhao, Z., Li, Y. N., Song, Z. G., & Chang, H. L. (2022). Application of Digital PCR in the Detection of Genetically Modified Plants. Chinese Journal of Biological Control, 38(5), 1143–1148. https://doi.org/10.16409/j.cnki.2095-039x.2022.09.012
[80]. Jiang, F., & Doudna, J. A. (2017). CRISPR-Cas9 Structures and Mechanisms. Annual Review of Biophysics, 46(1), 505–529. https://doi.org/10.1146/annurev-biophys-070316-033624
[81]. Wang, Q., Zhang, B., Xu, X., Long, F., & Wang, J. (2018). CRISPR-Typing PCR (ctPCR), a New Cas9-Based DNA Detection Method. Scientific Reports, 8(1), 1–10. https://doi.org/10.1038/s41598-018-32329-x
[82]. Perchet, T., Chea, S., Hasan, M., Cumano, A., & Golub, R. (2017). Single-cell Gene Expression Using Multiplex RT-qPCR to Characterize Heterogeneity of Rare Lymphoid Populations. Journal of Visualized Experiments (119), e54858. https://doi.org/10.3791/54858
[83]. Zhu, J. L., Liang, J. S., Zhang, P., Wang, W. W., Lin, T. S. Q., Xie, X. Y., Su, R., & Tang, W. (2019). A Rapid Detection Method for Potato Late Blight Caused by Phytophthora infestans Based on qPCR and LAMP Assays. Crops (6), 168–176. https://doi.org/10.16035/j.issn.1001-7283.2019.06.027
[84]. Jian, S., Hsiao, C., Chen, S., Weng, C., Kuo, T., Wu, D., et al. (2014). Utilization of Liquid Chromatography Mass Spectrometry Analyses to Identify LKB1–APC Interaction in Modulating Wnt/β-Catenin Pathway of Lung Cancer Cells. Molecular Cancer Research, 12(4), 622–635. https://doi.org/10.1158/1541-7786.MCR-13-0496
[85]. Fairbairn, D. W., Olive, P. L., & O'Neill, K. L. (1995). The comet assay: a comprehensive review. Mutation Research-Fundamental and Molecular Mechanisms of Mutagenesis, 339(1), 37–59. https://doi.org/10.1016/0027-5107(95)00006-9
[86]. Gomez Martinez, A. E., Lam, T., & Herr, A. E. (2025). Paired Analyses of Nuclear Protein Targets and Genomic DNA by Single-Cell Western Blot and Single-Cell PCR. bioRxiv. https://doi.org/10.1101/2025.01.01.489047
[87]. Sayour, N. V., Tóth, V. É., Nagy, R. N., Vörös, I., Gergely, T. G., Onódi, Z., Nagy, N., Bödör, C., Váradi, B., Ruppert, M., Radovits, T., Bleckwedel, F., Zelarayán, L. C., Pacher, P., Ágg, B., Görbe, A., Ferdinandy, P., & Varga, Z. V. (2023). Droplet Digital PCR Is a Novel Screening Method Identifying Potential Cardiac G-Protein-Coupled Receptors as Candidate Pharmacological Targets in a Rat Model of Pressure-Overload-Induced Cardiac Dysfunction. International Journal of Molecular Sciences, 24(18), 13826. https://doi.org/10.3390/ijms241813826
Cite this article
Zhu,Z. (2025). Breakthroughs and Applications of Emerging Protein and Gene Detection Technologies. Journal of Clinical Technology and Theory,3(2),52-65.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Journal:Journal of Clinical Technology and Theory
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Moritz, C. P. (2020). 40 years Western Blotting: A Scientific Birthday Toast. Journal of Proteomics, 212, 103575. https://doi.org/10.1016/j.jprot.2019.103571
[2]. Begum, H., Murugesan, P., & Tangutur, A. D. (2022). Western Blotting: a Powerful Staple in Scientific and Biomedical Research. Biotechniques, 73(1), 58–69. https://doi.org/10.2144/btn-2022-0003
[3]. Singh, K. K., Gupta, A., Bharti, C., & Sharma, H. (2021). Emerging Techniques of Western Blotting for Purification and Analysis of Protein. Future Journal of Pharmaceutical Sciences, 7(1). https://doi.org/10.1186/s43094-021-00386-1
[4]. Yang, P., & Mahmood, T. (2012). Western blot: Technique, Theory, and Trouble Shooting. North American Journal of Medical Sciences, 4(9), 429. https://doi.org/10.4103/1947-2714.100998
[5]. Towbin, H., Staehelin, T., & Gordon, J. (1979). Electrophoretic Transfer of Proteins from Polyacrylamide Gels to Nitrocellulose Sheets: Procedure and Some Applications. Proceedings of the National Academy of Sciences of the United States of America, 76(9), 4350–4354. https://doi.org/10.1073/pnas.76.9.4350
[6]. Litovchick, L. (2018). Immunoblotting: Transfer of Proteins from Gels to Membranes. Cold Spring Harbor Protocols, 2018(10), pdb.prot98442. https://doi.org/10.1101/pdb.prot098442
[7]. Sule, R., Rivera, G., & Gomes, A. V. (2023). Western Blotting (Immunoblotting): History, Theory, Uses, Protocol and Problems. Biotechniques, 75(3), 99–114. https://doi.org/10.2144/btn-2022-0034
[8]. Heidebrecht, F., Heidebrecht, A., Schulz, I., Behrens, S. E., & Bader, A. (2009). Improved Semiquantitative Western Blot Technique with Increased Quantification Range. Journal of Immunological Methods, 345(1–2), 40–48. https://doi.org/10.1016/j.jim.2009.02.005
[9]. Bose, U., Wijffels, G., Howitt, C. A., & Colgrave, M. L. (2019). Proteomics: Tools of the Trade. In J. L. Capelo-Martínez (Ed.), Emerging Sample Treatments in Proteomics (pp. 1–22). Springer. https://doi.org/10.1007/978-3-030-12298-0_1
[10]. Han, X. L., Yang, J. R., Bao, F. K., & Liu, A. H. (2017). Progress of Stain-Free Technology and Total Protein Analysis in Western Blotting. Letters in Biotechnology, 28(5), 709–714. https://doi.org/10.3969/j.issn.1009-0002.2017.05.028
[11]. Lallier, S. W., Hill, C. L., Nichols, D. P., & Reynolds, S. D. (2019). Protein Abundance Determination: An Optimized Western Blot Workflow. Annals of Clinical and Laboratory Science, 49(4), 507–512.
[12]. Kong, W., Li, Y., Cheng, S., Yan, C., An, S., Dong, Z., Yan, L., & Yuan, Y. (2016). Luminex xMAP Combined with Western Blot Improves HIV Diagnostic Sensitivity. Journal of Virological Methods, 227, 1–5. https://doi.org/10.1016/j.jviromet.2015.10.019
[13]. Mullis, K., Faloona, F., Scharf, S., Saiki, R., Horn, G., & Erlich, H. (1986). Specific Enzymatic Amplification of DNA in Vitro: the Polymerase Chain Reaction. Cold Spring Harbor Symposia on Quantitative Biology, 51(Pt 1), 263–273. https://doi.org/10.1101/sqb.1986.051.01.032
[14]. Smith, C. J., & Osborn, A. M. (2009). Advantages and Limitations of Quantitative PCR (Q-PCR)-based Approaches in Microbial Ecology. FEMS Microbiology Ecology, 67(1), 6–20. https://doi.org/10.1111/j.1574-6941.2008.00629.x
[15]. Tajadini, M., Panjehpour, M., & Javanmard, S. (2014). Comparison of SYBR Green and TaqMan Methods in Quantitative Real-time Polymerase Chain Reaction Analysis of Four Adenosine Receptor Subtypes. Advanced Biomedical Research, 3(1), 85. https://doi.org/10.4103/2277-9175.127998
[16]. Li, B., & Yan, T. (2021). Next Generation Sequencing Reveals Limitation of QPCR Methods in Quantifying Emerging Antibiotic Resistance Genes (ARGs) in the Environment. Applied Microbiology and Biotechnology, 105(7), 2925–2936. https://doi.org/10.1007/s00253-021-11202-6
[17]. Zhao, K., Shi, K., Zhou, Q., Xiong, C., Mo, S., Zhou, H., Long, F., Wei, H., Hu, L., & Mo, M. (2022). The Development of a Multiplex Real-Time Quantitative PCR Assay for the Differential Detection of the Wild-Type Strain and the MGF505-2R, EP402R and I177L Gene-Deleted Strain of the African Swine Fever Virus. Animals, 12(14), 1754. https://doi.org/10.3390/ani12141754
[18]. Zhang, Q., Wang, J., Deng, F., Yan, Z., Xia, Y., Wang, Z., Ye, J., Deng, Y., Zhang, Z., Qiao, M., Li, R., Denduluri, S. K., Wei, Q., Zhao, L., Lu, S., Wang, X., Tang, S., Liu, H., Luu, H. H., Haydon, R. C., He, T. C., & Jiang, L. (2015). TqPCR: A Touchdown qPCR Assay with Significantly Improved Detection Sensitivity and Amplification Efficiency of SYBR Green qPCR. PLOS ONE, 10(7), e0132666. https://doi.org/10.1371/journal.pone.0132666
[19]. Hu, X., Zang, X., & Lv, Y. (2021). Detection of Circulating Tumor Cells: Advances and Critical Concerns. Oncology Letters, 21(5), 422. https://doi.org/10.3892/ol.2021.12992
[20]. Hermann-Bank, M. L., Skovgaard, K., Stockmarr, A., Larsen, N., & Molbak, L. (2013). The Gut Microbiotassay: a High-Throughput QPCR Approach Combinable with Next Generation Sequencing to Study Gut Microbial Diversity. BMC Genomics, 14, 788. https://doi.org/10.1186/1471-2164-14-788
[21]. Logan, J. M. J., & Edwards, K. J. (2004). An Overview of Real-Time PCR Platforms. https://doi.org/10.1007/978-1-4020-2400-9_2
[22]. Kang, C., Yamauchi, K. A., Vlassakis, J., Sinkala, E., Duncombe, T. A., & Herr, A. E. (2016). Single Cell–Resolution Western Blotting. Nature Protocols, 11(8), 1508–1530. https://doi.org/10.1038/nprot.2016.089
[23]. Hughes, A. J., Spelke, D. P., Xu, Z., Kang, C., Schaffer, D. V., & Herr, A. E. (2014). Single-cell Western Blotting. Nature Methods, 11(7), 749–755. https://doi.org/10.1038/nmeth.2992
[24]. Kang, C., Lin, J. G., Xu, Z., Kumar, S., & Herr, A. E. (2014). Single-Cell Western Blotting after Whole-Cell Imaging to Assess Cancer Chemotherapeutic Response. Analytical Chemistry, 86(20), 10429–10436. https://doi.org/10.1021/ac502932t
[25]. Yamauchi, K. A., & Herr, A. E. (2017). Subcellular Western Blotting of Single Cells. Microsystems & Nanoengineering, 3(1), 1–10. https://doi.org/10.1038/micronano.2016.79
[26]. Li, J. J., Bickel, P. J., & Biggin, M. D. (2014). System Wide Analyses Have Underestimated Protein Abundances and the Importance of Transcription in Mammals. PeerJ, 2, e270. https://doi.org/10.7717/peerj.270
[27]. Durmus, N. G., Tekin, H. C., Guven, S., Sridhar, K., Arslan Yildiz, A., Calibasi, G., Ghiran, I., Davis, R. W., Steinmetz, L. M., & Demirci, U. (2015). Magnetic Levitation of Single Cells. Proceedings of the National Academy of Sciences of the United States of America, 112(28), 8606–8611. https://doi.org/10.1073/pnas.1509250112
[28]. Karin, M., & Greten, F. R. (2005). NF-κB: Linking Inflammation and Immunity to Cancer Development and Progression. Nature Reviews Immunology, 5(10), 749–759. https://doi.org/10.1038/nri1703
[29]. Zeder-Lutz, G., Cherouati, N., Reinhart, C., Pattus, F., & Wagner, R. (2006). Dot-Blot Immunodetection as a Versatile and High-Throughput Assay to Evaluate Recombinant GPCRs Produced in the Yeast Pichia Pastoris. Protein Expression and Purification, 50(1), 118–127. https://doi.org/10.1016/j.pep.2006.03.013
[30]. Ortega Ibarra, J. M., Cifuentes-Castro, V. H., Medina-Ceja, L., & Morales-Villagrán, A. (2021). Nano Dot Blot: An Alternative Technique for Protein Identification and Quantification in a High Throughput Format. Journal of Neuroscience Methods, 358, 109194. https://doi.org/10.1016/j.jneumeth.2021.109194
[31]. Klabunde, T., & Hessler, G. (2002). Drug Design Strategies for Targeting G-protein-coupled Receptors. ChemBioChem, 3(10), 928–944. https://doi.org/10.1002/1439-7633(20021004)3:10<928::AID-CBIC928>3.0.CO;2-5
[32]. Scholl, B., Liu, H. Y., Long, B. R., McCarty, O. J., O'Hare, T., Druker, B. J., & Vu, T. Q. (2009). Single Particle Quantum Dot Imaging Achieves Ultrasensitive Detection Capabilities for Western Immunoblot Analysis. ACS Nano, 3(6), 1318–1328. https://doi.org/10.1021/nn900390t
[33]. Bakalova, R., Zhelev, Z., Ohba, H., & Baba, Y. (2005). Quantum Dot-Based Western Blot Technology for Ultrasensitive Detection of Tracer Proteins. Journal of the American Chemical Society, 127(26), 9328–9329. https://doi.org/10.1021/ja0510055
[34]. Wald, F. V. (1977). Applications of CdTe. A Review. Revue de Physique Appliquée, 12(2), 277–290. https://doi.org/10.1051/rphysap:01977001202027700
[35]. Chen, W., Xu, D., Liu, L., Peng, C., Zhu, Y., Ma, W., Bian, A., Li, Z., Jin, Z., Zhu, S. (2009). Ultrasensitive Detection of Trace Protein by Western Blot Based on POLY-Quantum Dot Probes. Analytical Chemistry, 81(21), 9194–9198. https://doi.org/10.1021/ac901429a
[36]. Allen, S. J., & Dawbarn, D. (2006). Clinical Relevance of the Neurotrophins and Their Receptors. Clinical Science, 110(2), 175–191. https://doi.org/10.1042/CS20050347
[37]. Su, X. L., & Li, Y. (2004). Quantum Dot Biolabeling Coupled with Immunomagnetic Separation for Detection of Escherichia Coli O157:H7. Analytical Chemistry, 76(16), 4806–4810. https://doi.org/10.1021/ac049513k
[38]. Li, H., & Han, C. (2008). Sonochemical Synthesis of Cyclodextrin-Coated Quantum Dots for Optical Detection of Pollutant Phenols in Water. Chemistry of Materials, 20(19), 6053–6059. https://doi.org/10.1021/cm8009176
[39]. Wang, L., Liu, S., Peng, J., & He, Y. (2010). The Synthesis of Mono-6-Thio-β-Cyclodextrin Capped CdTe QDs and Its Interaction with Neutral Red. Science China Chemistry, 53(6), 1358–1365. https://doi.org/10.1007/s11426-010-3092-2
[40]. Li, H., & Qu, F. (2007). Synthesis of CdTe Quantum Dots in Sol−Gel-Derived Composite Silica Spheres Coated with Calix[4]arene as Luminescent Probes for Pesticides. Chemistry of Materials, 19(17), 4148–4154. https://doi.org/10.1021/cm0700089
[41]. Duchesne, L., & Fernig, D. G. (2007). Silver and Gold Nanoparticle-Coated Membranes for Femtomole Detection of Small Proteins and Peptides by Dot and Western Blot. Analytical Biochemistry, 362(2), 287–289. https://doi.org/10.1016/j.ab.2006.09.014
[42]. Emami, T., Madani, R., Rezayat, S. M., Golchinfar, F., & Sarkar, S. (2012). Applying of Gold Nanoparticle to Avoid Diffusion of the Conserved Peptide of Avian Influenza Nonstructural Protein from Membrane in Western Blot. Journal of Applied Poultry Research, 21(3), 563–566. https://doi.org/10.3382/japr.2011-00343
[43]. Shen, J., Hu, Y., Shi, M., Li, N., Ma, H., & Ye, M. (2010). One Step Synthesis of Graphene Oxide−Magnetic Nanoparticle Composite. The Journal of Physical Chemistry C, 114(3), 1498–1503. https://doi.org/10.1021/jp909756r
[44]. Green, A. A., & Hersam, M. C. (2010). Emerging Methods for Producing Monodisperse Graphene Dispersions. The Journal of Physical Chemistry Letters, 1(2), 544–549. https://doi.org/10.1021/jz900235f
[45]. Lin, H., Huo, J., Zhang, A., Liu, Y., Wang, Q., Cai, Y., Ying, W., Qin, W., Zhang, Y., & Qian, X. (2012). A Sensitive Dual Signal Amplification Method for Western Blotting Based On Antibody-Functionalised Graphene Oxide and Gold Nanoparticles. Analyst, 137(16), 3620–3623. https://doi.org/10.1039/C2AN36200A
[46]. Wu, D., Qin, J., & Lin, B. (2008). Electrophoretic Separations on Microfluidic Chips. Journal of Chromatography A, 1184(1–2), 542–559. https://doi.org/10.1016/j.chroma.2007.12.016
[47]. Whitesides, G. M. (2006). The Origins and The Future of Microfluidics. Nature, 442(7101), 368–373. https://doi.org/10.1038/nature05058
[48]. Kurien, B. T., & Scofield, R. H. (Eds.). (2015). Western Blotting: Methods and Protocols (Methods in Molecular Biology, Vol. 1312). Humana Press.
[49]. Sanders, B. J., Kim, D. C., & Dunn, R. C. (2016). Recent Advances in Microscale Western Blotting. Analytical Methods, 8(39), 7002–7013. https://doi.org/10.1039/C6AY01947A
[50]. Jin, S., Furtaw, M. D., Chen, H., Lamb, D. T., Ferguson, S. A., Arvin, N. E., Dawod, M., & Kennedy, R. T. (2016). Multiplexed Western Blotting Using Microchip Electrophoresis. Analytical Chemistry, 88(13), 6703–6710. https://doi.org/10.1021/acs.analchem.6b00705
[51]. Hughes, A. J., & Herr, A. E. (2012). Microfluidic Western blotting. Proceedings of the National Academy of Sciences of the United States of America, 109(52), 21450–21455. https://doi.org/10.1073/pnas.1214643109
[52]. Ramautar, R., Demirci, A., & Jong, G. J. D. (2006). Capillary Electrophoresis in Metabolomics. TrAC Trends in Analytical Chemistry, 25(5), 455–466. https://doi.org/10.1016/j.trac.2006.03.003
[53]. Mishra, M., Tiwari, S., & Gomes, A. V. (2017). Protein Purification and Analysis: Next Generation Western Blotting Techniques. Expert Review of Proteomics, 14(11), 1037–1053. https://doi.org/10.1080/14789450.2017.1388167
[54]. Franciosi, L., Govorukhina, N., Fusetti, F., Poolman, B., Lodewijk, M. E., Timens, W., Postma, D., ten Hacken, N., & Bischoff, R. (2013). Proteomic Analysis of Human Epithelial Lining Fluid by Microfluidics‐Based NanoLC‐MS/MS: A Feasibility Study. Electrophoresis, 34(18), 2683–2694. https://doi.org/10.1002/elps.201300020
[55]. Wang, Y., Guan, Z. Y., Shi, S. W., Jiang, Y. R., Zhang, J., Yang, Y., Wu, Q., Wu, J., Chen, J. B., Ying, W. X., Xu, Q. Q., Fan, Q. X., Wang, H. F., Zhou, L., Wang, L., Fang, J., Pan, J. Z., & Fang, Q. (2024). Pick-up Single-Cell Proteomic Analysis for Quantifying up to 3000 Proteins in a Mammalian Cell. Nature Communications, 15(1), 1–13. https://doi.org/10.1038/s41467-024-45659-4
[56]. Creamer, J. S., Oborny, N. J., & Lunte, S. M. (n.d.). Recent Advances in the Analysis of Therapeutic Proteins by Capillary and Microchip Electrophoresis. Analytical Methods. https://doi.org/10.1039/C7AY00841B
[57]. Vollmer, F., & Arnold, S. (2008). Whispering-Gallery-Mode Biosensing: Label-Free Detection Down to Single Molecules. Nature Methods, 5(7), 591–596. https://doi.org/10.1038/nmeth.1221
[58]. Hunt, H. K., & Armani, A. M. (2010). Label-Free Biological and Chemical Sensors. Nanoscale, 2(9), 1544. https://doi.org/10.1039/C0NR00201A
[59]. Wang, Z., Zhou, B., & Zhang, A. P. (2024). High-Q WGM Microcavity-Based Optofluidic Sensor Technologies for Biological Analysis. Biomicrofluidics, 18(4), 041502. https://doi.org/10.1063/5.0175538
[60]. Arnold, S., Ramjit, R., Keng, D., Kolchenko, V., & Teraoka, I. (2008). Micro Particle Photophysics Illuminates Viral Bio-sensing. Faraday Discussions, 137, 65–83. https://doi.org/10.1039/B702920A
[61]. He, S., Zhang, Y., Wang, P., Xu, X., Zhu, K., Pan, W., Liu, W., Cai, K., Sun, J., Zhang, W., & Jiang, X. (2015). Multiplexed Microfluidic Blotting of Proteins and Nucleic Acids by Parallel, Serpentine Microchannels. Lab on a Chip, 15(1), 105–112. https://doi.org/10.1039/C4LC01167J
[62]. Jensen, B. C., Swigart, P. M., & Simpson, P. C. (2009). Ten Commercial Antibodies for Alpha-1-Adrenergic Receptor Subtypes are Nonspecific. Naunyn-Schmiedeberg's Archives of Pharmacology, 379(4), 409–412. https://doi.org/10.1007/s00210-008-0368-6
[63]. Michel, M. C., Wieland, T., & Tsujimoto, G. (2009). How Reliable are G-Protein-Coupled Receptor Antibodies? Naunyn-Schmiedeberg's Archives of Pharmacology, 379(4), 385–388. https://doi.org/10.1007/s00210-009-0395-y
[64]. Surappa, S., Multani, P., Parlatan, U., Sinawang, P. D., Kaifi, J., Akin, D., & Demirci, U. (2024). Integrated “Lab-on-a-Chip” Microfluidic Systems for Isolation, Enrichment, and Analysis of Cancer Biomarkers. Lab Chip, 23(13), 2942–2958. https://doi.org/10.1039/D4LC00314K
[65]. Fang, T., Peng, L., Yang, T., Cai, Q., Li, H., Li, H., & Cai, L. (2025). Development and Evaluation of Multiplex Real-Time PCR for Rapid Identifying Major Pathogenic Mycobacteria from Pulmonary and Extrapulmonary Clinical Samples in Eastern China. Heliyon, 11(1), e41384. https://doi.org/10.1016/j.heliyon.2024.e41384
[66]. Marras, S. A. E., Tyagi, S., Antson, D., & Kramer, F. R. (2019). Color-Coded Molecular Beacons for Multiplex PCR Screening Assays. PLoS One, 14(3), e213906. https://doi.org/10.1371/journal.pone.0213906
[67]. Lin, C., Yeh, K., Chang, Y., Hsu, N. C., & Chang, J. (2010). Rapid Detection of Epidermal Growth Factor Receptor Mutations with Multiplex PCR and Primer Extension in Lung Cancer. Journal of Biomedical Science, 17(1), 37. https://doi.org/10.1186/1423-0127-17-37
[68]. Hou, D., Chen, C., Seely, E. J., Chen, S., & Song, Y. (2016). High-Throughput Sequencing-Based Immune Repertoire Study during Infectious Disease. Frontiers in Immunology, 7, 336. https://doi.org/10.3389/fimmu.2016.00336
[69]. Van der Heyden, H., Wallon, T., Lévesque, C. A., & Carisse, O. (2019). Detection and Quantification of Pythium tracheiphilum in Soil by Multiplex Real-Time qPCR. Plant Disease, 103(3), 475–483. https://doi.org/10.1094/PDIS-03-18-0419-RE
[70]. Eckford-Soper, L. K., & Daugbjerg, N. (2015). Development of a Multiplex Real-Time qPCR Assay for Simultaneous Enumeration of up to Four Marine Toxic Bloom-Forming Microalgal Species. Harmful Algae, 48, 37–43. https://doi.org/10.1016/j.hal.2015.05.006
[71]. Lin, J., Su, G., Su, W., & Zhou, C. (2017). Progress in digital PCR technology and application. Chinese Journal of Biotechnology, 33(2), 170–177. https://doi.org/10.13345/j.cjb.160269
[72]. Postel, M., Roosen, A., Laurent-Puig, P., Taly, V., & Wang-Renault, S. (2018). Droplet-Based Digital PCR and Next Generation Sequencing for Monitoring Circulating Tumor DNA: A Cancer Diagnostic Perspective. Expert Review of Molecular Diagnostics, 18(1), 7–17. https://doi.org/10.1080/14737159.2017.1394582
[73]. Pawinwongchai, J. (2020). Study of Platelet Production from Megakaryocyte by Using Induced Pluripotent Stem Cell. Chulalongkorn University Theses and Dissertations, 442. https://digital.car.chula.ac.th/chulaetd/442
[74]. Qu, Y. L. (2019). Research on the Detection of Salivary Exosome Tumor Markers by Digital PCR Chip [Master’s thesis]. Dalian Medical University.
[75]. Renault, C., Bolloré, K., Pisoni, A., Motto-Ros, C., Van de Perre, P., Reynes, J., & Tuaillon, E. (2022). Accuracy of Real-Time PCR and Digital PCR for the Monitoring of Total HIV DNA under Prolonged Antiretroviral Therapy. Scientific Reports, 12(1), 1–10. https://doi.org/10.1038/s41598-022-13581-8
[76]. Banada, P. P., Deshpande, S., Banik, S., Shah, D., Koshy, R., Patel, B., Kwiatkowski, R., Persing, D., & Alland, D. (2019). Multiplex Detection of Three Select Agents Directly from Blood by Use of the GeneXpert System. Journal of Clinical Microbiology, 57(5), e00036-19. https://doi.org/10.1128/JCM.00036-19
[77]. Bai, H., Liu, Y., Gao, L., Wang, T., Zhang, X., Hu, J., Ding, L., Zhang, Y., Wang, Q., Wang, L., Li, J., Zhang, Z., Wang, Y., Shen, C., Ying, B., Niu, X., & Hu, W. (2024). A portable All-In-One Microfluidic Device with Real-Time Colorimetric LAMP for HPV16 and HPV18 DNA Point-of-Care Testing. Biosensors and Bioelectronics, 248, 115968. https://doi.org/10.1016/j.bios.2023.115968
[78]. Olwagen, C. P., Adrian, P. V., & Madhi, S. A. (2019). Performance of the Biomark HD Real-Time qPCR System (Fluidigm) for the Detection of Nasopharyngeal Bacterial Pathogens and Streptococcus Pneumoniae Typing. Scientific Reports, 9(1), 1–10. https://doi.org/10.1038/s41598-019-42846-y
[79]. You, Y. H., Zhao, Z., Li, Y. N., Song, Z. G., & Chang, H. L. (2022). Application of Digital PCR in the Detection of Genetically Modified Plants. Chinese Journal of Biological Control, 38(5), 1143–1148. https://doi.org/10.16409/j.cnki.2095-039x.2022.09.012
[80]. Jiang, F., & Doudna, J. A. (2017). CRISPR-Cas9 Structures and Mechanisms. Annual Review of Biophysics, 46(1), 505–529. https://doi.org/10.1146/annurev-biophys-070316-033624
[81]. Wang, Q., Zhang, B., Xu, X., Long, F., & Wang, J. (2018). CRISPR-Typing PCR (ctPCR), a New Cas9-Based DNA Detection Method. Scientific Reports, 8(1), 1–10. https://doi.org/10.1038/s41598-018-32329-x
[82]. Perchet, T., Chea, S., Hasan, M., Cumano, A., & Golub, R. (2017). Single-cell Gene Expression Using Multiplex RT-qPCR to Characterize Heterogeneity of Rare Lymphoid Populations. Journal of Visualized Experiments (119), e54858. https://doi.org/10.3791/54858
[83]. Zhu, J. L., Liang, J. S., Zhang, P., Wang, W. W., Lin, T. S. Q., Xie, X. Y., Su, R., & Tang, W. (2019). A Rapid Detection Method for Potato Late Blight Caused by Phytophthora infestans Based on qPCR and LAMP Assays. Crops (6), 168–176. https://doi.org/10.16035/j.issn.1001-7283.2019.06.027
[84]. Jian, S., Hsiao, C., Chen, S., Weng, C., Kuo, T., Wu, D., et al. (2014). Utilization of Liquid Chromatography Mass Spectrometry Analyses to Identify LKB1–APC Interaction in Modulating Wnt/β-Catenin Pathway of Lung Cancer Cells. Molecular Cancer Research, 12(4), 622–635. https://doi.org/10.1158/1541-7786.MCR-13-0496
[85]. Fairbairn, D. W., Olive, P. L., & O'Neill, K. L. (1995). The comet assay: a comprehensive review. Mutation Research-Fundamental and Molecular Mechanisms of Mutagenesis, 339(1), 37–59. https://doi.org/10.1016/0027-5107(95)00006-9
[86]. Gomez Martinez, A. E., Lam, T., & Herr, A. E. (2025). Paired Analyses of Nuclear Protein Targets and Genomic DNA by Single-Cell Western Blot and Single-Cell PCR. bioRxiv. https://doi.org/10.1101/2025.01.01.489047
[87]. Sayour, N. V., Tóth, V. É., Nagy, R. N., Vörös, I., Gergely, T. G., Onódi, Z., Nagy, N., Bödör, C., Váradi, B., Ruppert, M., Radovits, T., Bleckwedel, F., Zelarayán, L. C., Pacher, P., Ágg, B., Görbe, A., Ferdinandy, P., & Varga, Z. V. (2023). Droplet Digital PCR Is a Novel Screening Method Identifying Potential Cardiac G-Protein-Coupled Receptors as Candidate Pharmacological Targets in a Rat Model of Pressure-Overload-Induced Cardiac Dysfunction. International Journal of Molecular Sciences, 24(18), 13826. https://doi.org/10.3390/ijms241813826









