1. Introduction
High blood sugar levels are the main cause of diabetes, a chronic condition that affects millions of people. The 10th edition of the IDF's Diabetes Atlas 2021, released on the official website of the International Diabetes Federation (IDF) (www.diabetalas.org), states that 537 million people are currently suffering from the condition. By 2045, it is expected that the number of people with this disease will reach over 800 million[1]. This underscores the need to enhance the precision of diagnosis and provide prompt and comprehensive care for all individuals with diabetes, especially those who are unaware of their condition[2].
Individuals who are suffering from diabetes have a higher chance of developing various complications. These include kidney failure, heart disease, blindness, and nerve damage[3]. Diabetic eye disease refers to the group of eye conditions that can result from diabetes. These include: diabetic retinopathy, macular edema, glaucoma, cataracts[4]. The most common eye condition is diabetic retinopathy. Approximately 93 million individuals suffer from eye damage due to diabetes, and the leading cause of blindness among them is diabetic retinopathy (DR). DR damages the blood vessels located at the back of the eye[5], which can lead to vision loss if not treated.
Although managing diabetes and preventing eye damage can help avoid major health complications, it is still important to carry out screening for diabetic retinopathy. This can only be done through the help of a trained eye doctor. Unfortunately, it is not always feasible or efficient to perform this procedure on every individual. With the help of deep learning technology, a screening method can be developed that can accurately detect diabetic retinopathy[6].
2. Related Work
Early studies on the use of deep learning technology for detecting retinal diseases have been conducted. These studies utilized machine learning methods to classify a large amount of funds obtained from screening programs[7],[8],[9],[10]. Nevertheless, the extraction of features from this method is not ideal. It requires a lot of expertise to identify the exact features of a given dataset. Also, the time-consuming tasks involved in the extraction, recognition, and feature selection process make it impractical for large-scale applications.
In the field of classification and detection of retinal diseases, deep learning (DL) has been widely used. Unlike machine learning, its methods do not require the use of hand-crafted features extraction. A Google research team has developed an advanced model that can diagnose diabetes mellitus retinopathy (DMR)[11]. Several studies on retinal image classification opted for binary classification to address issues related to distinguishing between normal cases and a specific disease. In order to classify diabetic retinopathy in the Messidor dataset, Lam and his team used pre-trained networks (GoogleNet and AlexNet). They utilized selective contrast-limited adaptive histogram equalization (CLAHE) and achieved a significant improvement in their performance when it came to identifying subtle features[12]. A study conducted by Choi and his team revealed that a multi-class DL algorithm was able to automatically detect ten retinal disorders[13].
Some researchers have attained progress in the field of automatic detection of Diabetic Retinopathy (DR). However, challenges such as inadequate retinal image quality, unclear grading of retinal lesions, severe imbalances in category distribution, and a prevalent and challenging issue—insufficient images of DR lesions—require attention and resolution[14].
Transfer learning stands out as a potent research approach in the data science community, enabling the refinement of existing model algorithms for application in novel domains or tasks [15]. Rather than initiating the training of an entirely new model, transfer learning enables the utilization of a proficient pre-trained model, trained on abundant, high-quality labeled data. This approach facilitates the construction of a new model better suited to fulfilling specific tasks[16]. It can selectively transfer different pre-trained layers to the target task based on the characteristics of CNN classification tasks. Based on the above strengths, transfer learning is particularly suitable for deep learning research that is highly specialized but has a small dataset size, which can significantly reduce the dependence of model construction on data volume.
3. Dataset
3.1. Dataset Description
This experiment is based on the Diabetic Retinopathy 2015 Data Colored Resized database. All images have been resized and cropped, with the dimensions limited to a maximum size of 1024 pixels. For the 2015 Diabetic Retinopathy Detection images, the left and right eyes were collected from each subject. There are 35126 resized and cropped training set images and 53,576 resized and cropped test set images.
Clinicians have assessed each image, assigning a severity rating on a scale ranging from 0 to 4 for diabetic retinopathy:
level 0 - No Diabetic Retinopathy
level 1 - Mild diabetic retinopathy
level 2 - Moderate diabetic retinopathy
level 3 - Severe diabetic retinopathy
level 4 - Proliferative Diabetic Retinopathy
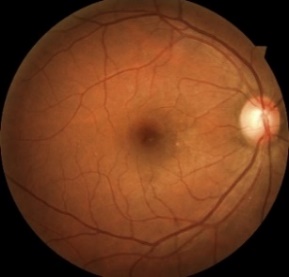
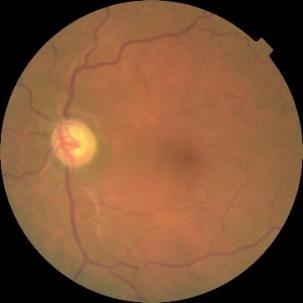
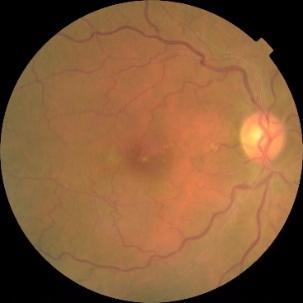
Figure 1. Level 0. Figure 2. Level 1. Figure 3. Level 2.
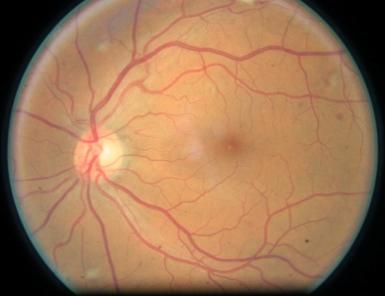
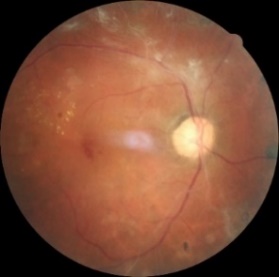
Figure 4. Level 3. Figure 5. Level 4.
3.2. Data Preprocessing
A random selection process selects 20% of the training dataset for validation. Training data is then categorized into five categories. This procedure is repeated for the testing and validation phases.
Because the pre-trained deep learning model that will be used in this experiment is based on the ImageNet, so, in this experiment, the average pixel intensity from each image channel will be subtracted by channel with reference to the ImageNet dataset. This ensures that the image intensities of the diabetic images have the same intensity range as the processed ImageNet images before training on the model.
The mean pixel intensities for the red, green, and blue channels in the ImageNet image are 103.939, 116.779, and 123.68, respectively[17]. The pre-trained model undergoes training by subtracting these averages from the image. Subtracting the mean values serves the purpose of normalizing the data features, with the goal of concentrating the data around 0 to mitigate issues like gradient vanishing and gradient explosion. This, in turn, facilitates faster convergence of the model. Furthermore, normalizing each channel ensures uniform gradient flow across all channels. Given the utilization of pre-trained models in this project, it is logical to normalize each channel in the same manner before feeding the images into the pre-trained network.
Besides, in order to expand the data, additional data is generated by performing affine transformations on the pixel coordinates of the image[18]. The main affine transformations are rotation, translation and scaling. Th research used horizontal and vertical flipping, which will produce images obtained by reflecting along the horizontal and vertical axes, respectively. Similarly, images are also translated by 10% pixels along the width and height directions, the rotation was limited to an angle of 20 degrees, and the scaling factor was defined within 0.8 to 1.2 of the original image size.
4. Methods
4.1. Model Construction
The article utilizes the Keras API. It is a powerful tool for developing deep learning models. It's written in Python and is based on various frameworks such as Theano, CNTK, and TensorFlow. Keras supports rapid experimentation, which allows for quick conversion of ideas into results. Therefore, it is a tool that can help us build deep learning models efficiently.
In this experiment, InceptionV3 model was utilized. InceptionV3 is a Google CNN. Instead of using a fixed-size convolutional filter at each layer, the InceptionV3 architecture uses different sized filters to extract features at different granularity levels. Besides, InceptionV3 is a deeper network with more initial layers. So, it can extract more accurate image features[19]. The convolutional block for the InceptionV3 layer is shown in figure 6.
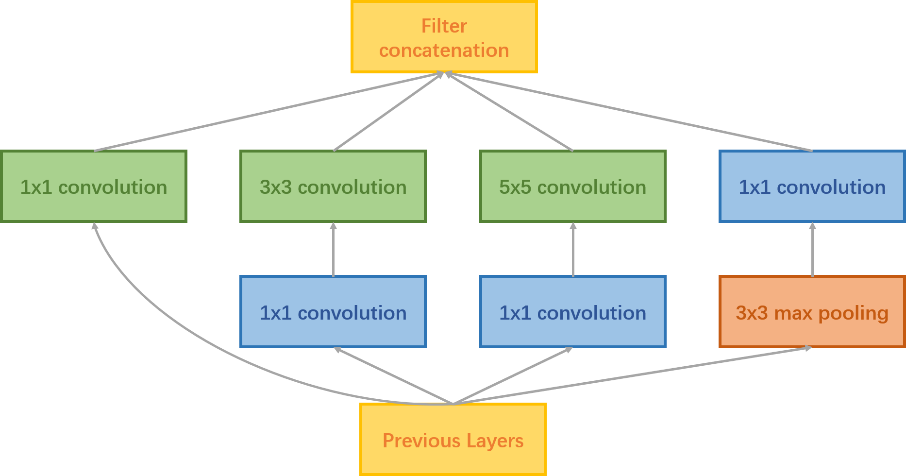
Figure 6. InceptionV3 Convolutional Block.
When training a model based on a small dataset, the weights of the entire network are more likely to be overfitted. However, freezing layers in transfer learning can reduce the number of weights that need to be trained, which can be considered as a kind of regularization, and the problem of overfitting can be reduced to some extent. So, it is important to find appropriate layers to freeze. Since the initial layers in a model learn generic features that are independent of the target domain, they are the most suitable layers for freezing. So, the first 30 layers of the network were frozen, which are mainly used to extract general features, which are basically similar to Gabor filters and color blobs.
Following the extraction of the output from the last maximum pooling layer of the pre-trained network, two additional modules are connected. These modules include a fully-connected layer with ReLU as the activation function and a dropout module. The dropout module aids in enhancing the model's generalization ability and improves the fitting capacity of neurons within the fully connected layer. To fulfill the classification task, a Softmax classifier was employed, connecting the fully connected layer of the InceptionV3 model to the LogSoftmax classifier. The depicted structure is illustrated in figure 7.
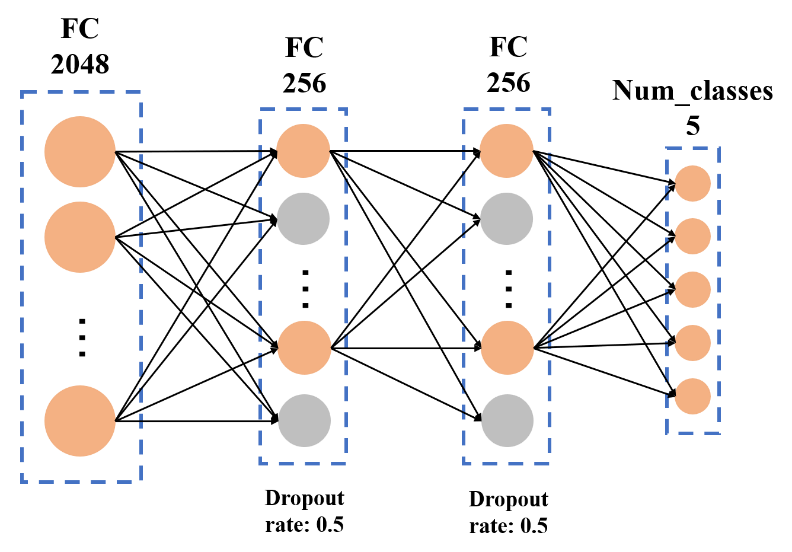
Figure 7. Fully Connected Layer.
4.2. Model Training
Classification of medical images is a challenging process due to the imbalance of class. Figure 8 shows the distribution of the training set's five categories.
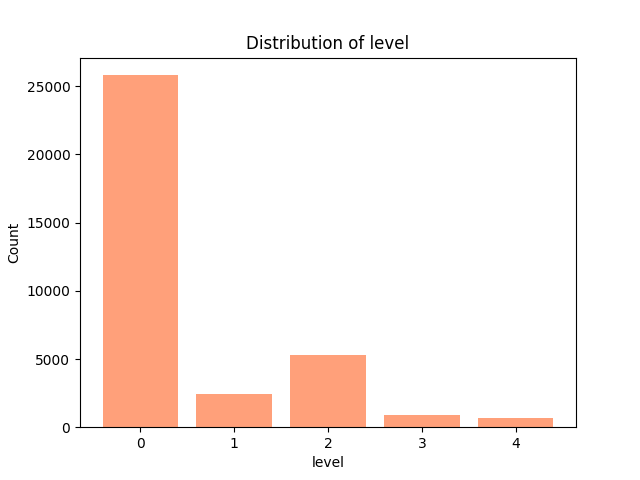
Figure 8. Distribution of Level.
Close to 73% of the training data falls into class 0, i.e. no diabetic retinopathy. Thus, if we happen to label all data as class 0, then the accuracy may be 73%. But in real life, we would rather misclassify a patient as having a problem (false-positive) when he or she does not actually have some kind of health problem than misclassify him or her as not having a problem (false-negative) when he or she does have some kind of health problem. Thus, even if the model learns to categorize all data as class 0, its 73% accuracy may not mean much. In order to tackle this problem, in the loss function, categories are assigned weights that are inversely proportional to their density[20]. This ensures that the loss function assigns a higher penalty to low-frequency categories when the model fails to categorize them correctly.
The corresponding weights for the different categories are shown in table 1.
Table 1. Weights for the different categories. |
Category of severity | Category weight |
Level 0 | 0.012035386 |
Level 1 | 0.127135056 |
Level 2 | 0.058696197 |
Level 3 | 0.364023421 |
Level 4 | 0.438197473 |
In order to measure the classification accuracy more precisely, the quadratic weighted kappa (QWK) statistic was also used to define the quality of the mode. The quadratic weighted kappa is defined as follows:
\( k = \frac{\sum _{i,j}{w_{i,j}}{O_{i,j}}}{\sum _{i,j}{w_{i,j}}{E_{i,j}}}\ \ \ (1) \)
The weights in the quadratic weighted kappa expression are defined as follows:
\( {w_{i,j}} = \frac{{(i-j)^{2}}}{N-1}\ \ \ (2) \)
Explanation of symbols in the above formula: N denotes the number of categories. \( {O_{i,j}} \) denotes the number of images that are predicted as category i and the actual category is j. \( {E_{i,j}} \) denotes the expected number of images that are predicted as category i and the actual category is j. It is assumed that the predicted category and the actual class are independent of each other.
In the training phase, the initial learning rate is established at 0.0001. Learning rate adjustments are made by monitoring losses on the validation set, with the learning rate decreasing at a multiplicative rate of 0.5. The optimizer employed to seek the optimal solution is Adam. Besides, small batches can be created dynamically using Keras to reduce the memory required for the training process. Dynamic batch generation is an efficient technique for creating small batches, and there are minimal performance issues when doing so. This is because methods such as Keras have a very efficient design.
5. Result
In this experiment, after training repetitively I found that when set the number of epochs to 50, training accuracy will become relative stable. Figure 9 shows the training loss during the training process and figure 10 shows the training accuracy and the validation accuracy. Finally, the InceptionV3 model gets close to 90.8% validation accuracy and a quadratic kappa score of 0.403.
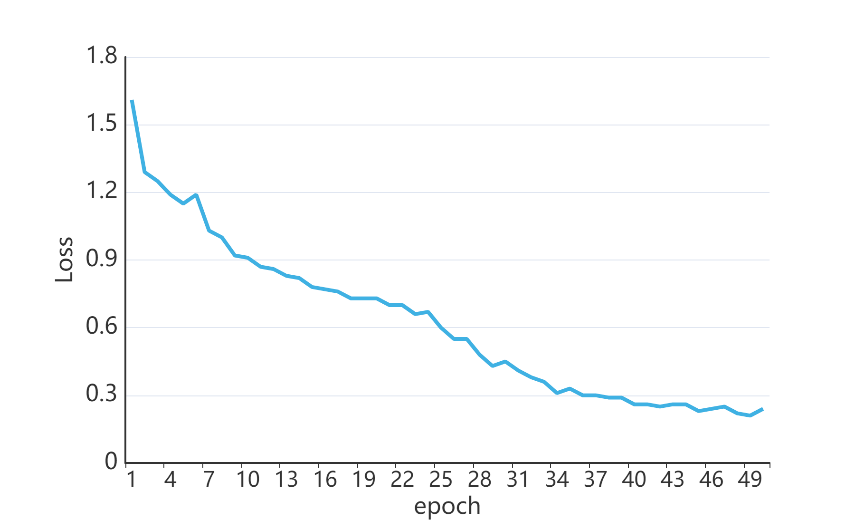
Figure 9. Training Loss.
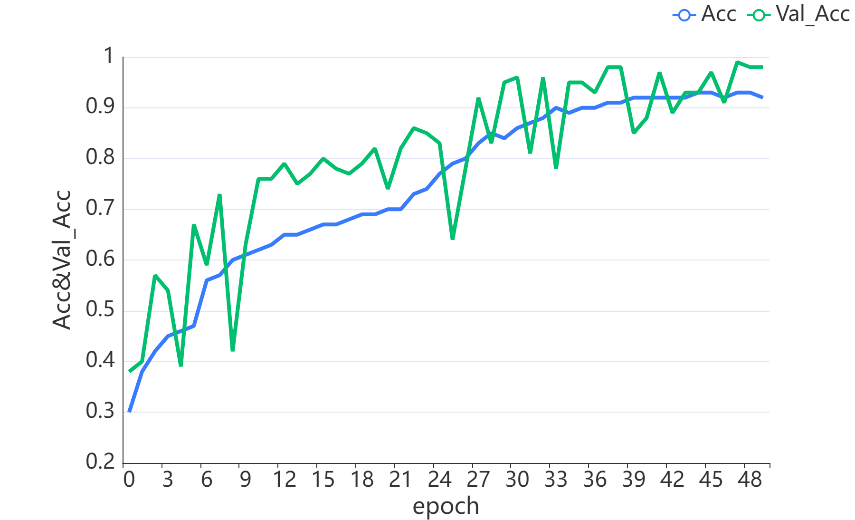
Figure 10. Training Accuracy and Validation Accuracy.
I tested the classifier model on a previously unseen dataset of 53,576 fundus images and classifier model resulted in an accuracy of 92.314%.
6. Conclusion
In this paper, transfer learning based on a pre-trained InceptionV3 model is implemented to classify DR into 5 classes and finally reaches the best accuracy 92.314%. After the pre-trained model, fully connected layers and classifiers are reconstructed to fit the specific task.
However, it is worth noting that the validation accuracy is not relatively stable during the training process and some- times there are large jumps in accuracy values. Considering that a portion of the neuron nodes have been discarded to circumvent the overfitting problem using dropout, the cause of the problem may stem from the data itself, such as overexposure or lighting problems and domain gap between different data acquisition equipment. This problem still needs to be addressed.
References
[1]. I. D. Federation. (2021) IDF Diabetes Atlas, 10th edn. Brussels, Belgium.
[2]. Ogurtsova K, Guariguata L, Barengo N C, et al. IDF diabetes Atlas: Global estimates of undiagnosed diabetes in adults for 2021[J]. Diabetes research and clinical practice, 2022, 183: 109118.
[3]. Diabetes complications. https://www.idf.org/aboutdiabetes/ complications.html.
[4]. M. Porta and J. Cunha-Vaz, Diabetes and the Eye. Cham: Springer International Publishing, 2019, pp. 1–43.
[5]. Sarhan A, Rokne J, Alhajj R, et al. Transfer learning through weighted loss function and group normalization for vessel segmentation from retinal images[C]//2020 25th International Conference on Pattern Recognition (ICPR). IEEE, 2021: 9211-9218.
[6]. Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs[J]. JAMA, 2016, 316(22): 2402-2410.
[7]. Wu J H, Liu T Y A, Hsu W T, et al. Performance and limitation of machine learning algorithms for diabetic retinopathy screening: meta-analysis[J]. Journal of medical Internet research, 2021, 23(7): e23863.
[8]. Bhatia K, Arora S, Tomar R. Diagnosis of diabetic retinopathy using machine learning classification algorithm[C]//2016 2nd international conference on next generation computing technologies (NGCT). IEEE, 2016: 347-351.
[9]. Tsao H Y, Chan P Y, Su E C Y. Predicting diabetic retinopathy and identifying interpretable biomedical features using machine learning algorithms[J]. BMC bioinformatics, 2018, 19: 111-121.
[10]. Reddy G T, Bhattacharya S, Ramakrishnan S S, et al. An ensemble based machine learning model for diabetic retinopathy classification[C]//2020 international conference on emerging trends in information technology and engineering (ic-ETITE). IEEE, 2020: 1-6.
[11]. Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs[J]. JAMA, 2016, 316(22): 2402-2410.
[12]. Sarki R, Ahmed K, Wang H, et al. Automated detection of mild and multi-class diabetic eye diseases using deep learning[J]. Health Information Science and Systems, 2020, 8(1): 32.
[13]. Choi J Y, Yoo T K, Seo J G, et al. Multi-categorical deep learning neural network to classify retinal images: A pilot study employing small database[J]. PloS one, 2017, 12(11): e0187336.
[14]. Qummar S, Khan F G, Shah S, et al. A deep learning ensemble approach for diabetic retinopathy detection[J]. IEEE Access, 2019, 7: 150530-150539.
[15]. Kandel I, Castelli M. Transfer learning with convolutional neural networks for diabetic retinopathy image classification. A review[J]. Applied Sciences, 2020, 10(6): 2021.
[16]. Li X, Pang T, Xiong B, et al. Convolutional neural networks based transfer learning for diabetic retinopathy fundus image classification[C]//2017 10th international congress on image and signal processing, biomedical engineering and informatics (CISP-BMEI). IEEE, 2017: 1-11.
[17]. Krizhevsky A, Sutskever I, Hinton G E. Imagenet classification with deep convolutional neural networks[J]. Advances in neural information processing systems, 2012, 25.
[18]. Mikołajczyk A, Grochowski M. Data augmentation for improving deep learning in image classification problem[C]//2018 international interdisciplinary PhD workshop (IIPhDW). IEEE, 2018: 117-122.
[19]. Xia X, Xu C, Nan B. Inception-v3 for flower classification[C]//2017 2nd international conference on image, vision and computing (ICIVC). IEEE, 2017: 783-787.
[20]. Bäck T, Fogel D B, Michalewicz Z. Handbook of evolutionary computation[J]. Release, 1997, 97(1): B1.
Cite this article
Zhang,L. (2024). Transfer learning approach for diabetic retinopathy detection using residual network. Applied and Computational Engineering,49,324-331.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 4th International Conference on Signal Processing and Machine Learning
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. I. D. Federation. (2021) IDF Diabetes Atlas, 10th edn. Brussels, Belgium.
[2]. Ogurtsova K, Guariguata L, Barengo N C, et al. IDF diabetes Atlas: Global estimates of undiagnosed diabetes in adults for 2021[J]. Diabetes research and clinical practice, 2022, 183: 109118.
[3]. Diabetes complications. https://www.idf.org/aboutdiabetes/ complications.html.
[4]. M. Porta and J. Cunha-Vaz, Diabetes and the Eye. Cham: Springer International Publishing, 2019, pp. 1–43.
[5]. Sarhan A, Rokne J, Alhajj R, et al. Transfer learning through weighted loss function and group normalization for vessel segmentation from retinal images[C]//2020 25th International Conference on Pattern Recognition (ICPR). IEEE, 2021: 9211-9218.
[6]. Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs[J]. JAMA, 2016, 316(22): 2402-2410.
[7]. Wu J H, Liu T Y A, Hsu W T, et al. Performance and limitation of machine learning algorithms for diabetic retinopathy screening: meta-analysis[J]. Journal of medical Internet research, 2021, 23(7): e23863.
[8]. Bhatia K, Arora S, Tomar R. Diagnosis of diabetic retinopathy using machine learning classification algorithm[C]//2016 2nd international conference on next generation computing technologies (NGCT). IEEE, 2016: 347-351.
[9]. Tsao H Y, Chan P Y, Su E C Y. Predicting diabetic retinopathy and identifying interpretable biomedical features using machine learning algorithms[J]. BMC bioinformatics, 2018, 19: 111-121.
[10]. Reddy G T, Bhattacharya S, Ramakrishnan S S, et al. An ensemble based machine learning model for diabetic retinopathy classification[C]//2020 international conference on emerging trends in information technology and engineering (ic-ETITE). IEEE, 2020: 1-6.
[11]. Gulshan V, Peng L, Coram M, et al. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs[J]. JAMA, 2016, 316(22): 2402-2410.
[12]. Sarki R, Ahmed K, Wang H, et al. Automated detection of mild and multi-class diabetic eye diseases using deep learning[J]. Health Information Science and Systems, 2020, 8(1): 32.
[13]. Choi J Y, Yoo T K, Seo J G, et al. Multi-categorical deep learning neural network to classify retinal images: A pilot study employing small database[J]. PloS one, 2017, 12(11): e0187336.
[14]. Qummar S, Khan F G, Shah S, et al. A deep learning ensemble approach for diabetic retinopathy detection[J]. IEEE Access, 2019, 7: 150530-150539.
[15]. Kandel I, Castelli M. Transfer learning with convolutional neural networks for diabetic retinopathy image classification. A review[J]. Applied Sciences, 2020, 10(6): 2021.
[16]. Li X, Pang T, Xiong B, et al. Convolutional neural networks based transfer learning for diabetic retinopathy fundus image classification[C]//2017 10th international congress on image and signal processing, biomedical engineering and informatics (CISP-BMEI). IEEE, 2017: 1-11.
[17]. Krizhevsky A, Sutskever I, Hinton G E. Imagenet classification with deep convolutional neural networks[J]. Advances in neural information processing systems, 2012, 25.
[18]. Mikołajczyk A, Grochowski M. Data augmentation for improving deep learning in image classification problem[C]//2018 international interdisciplinary PhD workshop (IIPhDW). IEEE, 2018: 117-122.
[19]. Xia X, Xu C, Nan B. Inception-v3 for flower classification[C]//2017 2nd international conference on image, vision and computing (ICIVC). IEEE, 2017: 783-787.
[20]. Bäck T, Fogel D B, Michalewicz Z. Handbook of evolutionary computation[J]. Release, 1997, 97(1): B1.









