1 Introduction
With the rapid development of genetic research and gene science and technology, gene editing technology has become one of the most promising technologies in this scientific and technological revolution. The team of He Jiankui from Southern University of Science and Technology in Shenzhen, China, suddenly announced the day before the second International Human Genome Editing Summit that a pair of gene-edited babies named Lulu and Nana had been born healthily in China in [1-3]November, which triggered a global academic shock after the news was sent. Gene editing technology not only has important applications in the field of medicine, but also plays a key role in ecology and environmental protection. By editing the genes of plants and microbes, researchers can create organisms that are more adaptable and resistant to stress to promote ecosystem restoration and conservation. For example, the use of gene editing technology to improve crops to grow in harsh environmental conditions can improve the stability and sustainability of agricultural production. Furthermore, it explores how the integration of artificial intelligence (AI) could confer additional advantages to gene editing practices. By synthesizing insights from both gene editing and AI domains, this paper aims to provide a comprehensive perspective on the convergence of these cutting-edge technologies and their potential synergies in shaping the future of bioengineering.
2 Related work
2.1 Explanation of CRISPR-Cas9 mechanism
Gene editing technology generally refers to the technology of modifying specific target genomic sites. In the 1980s, the first gene-editing technique, gene targeting (or gene knockout), was developed in mouse models [10]. In recent years, gene editing, GE technology is developing rapidly and is seen as a disruptive technology. [4] Gene editing techniques include morpholino oligomer,MO) [5]technology, Zinc finger nucleases (ZFNs) technology, transcriptional activator like effectant nucleases (TALENs) technology and clusters of regularly spaced short palindromic repeats (CRISPR) technology. CRISPR/Cas9 technology is the latest gene-editing technology, compared to ZFN technology and TALEN technology. The CRISPR/cas9 gene editing technology takes a different approach, relying on two parts: the Cas9 protein that cuts the target DNA, and the target site-specific guide RNA(RNA). In vitro synthesis of singleguide RNA(SgRNA) is designed according to the target DNA sequence that needs to be edited. The gene editing process is simply that the CRISPR/Cas9 system is guided by SgRNA to find the target sequence through matching, and then edited by Cas9 protein.
2.2 Principles of CRISPR/Cas9 gene editing
1) The highly variable spacer region of CRISPR is obtained
The acquisition of a highly variable interval region of CRISPR means that a short DNA sequence of the invading phage or plasmid DNA is integrated into the host bacterium's genome between two repeats at the CRISPR5' end, which can be divided into three steps:
Step 1: The proteins encoded by Cas1 and Cas2 scan the invading[5] DNA and identify the primary spacer sequence adjacent motifs (PAM), and then use the DNA sequence adjacent to PAM as a candidate primary spacer sequence.
Step 2: The Cas1/2 protein complex cuts the primary spacer sequence from the foreign DNA and inserts the primary spacer sequence downstream of the adjacent CRISPR sequence leader (an AT-rich region of 300-500bp length located upstream of the CRISPR gene).
Step 3: DNA is repaired via the nonhomologous end joining (NHEJ) or homologous directed repair (HDR) pathway, closing the open double-stranded notch. In this way, a new interval sequence is added to the CRISPR sequence of the genome.
2) Expression of CRISPR loci (including transcription and post-transcriptional maturation processing)
The CRISPR sequence is transcribed under the regulation of the precursor region to produce pre-crRNA and tracrRNA which is complementary to the pre-crRNA sequence. pre-crRNA forms sgRNA by complementary base pairing with tracrRNA and complex with Cas9 protein. It selects the corresponding spacer RNA according to the type of invader, and cuts the PAM with the help of RNase III, and finally forms a short crRNA. Most current gene editing using the CRISPR/Cas system uses synthetic Sgrnas.
3) Activation of CRISPR/Cas9 system (targeted interference)
The resulting complex of crRNA, tracrRNA, and Cas9 scans the entire foreign DNA sequence and identifies the original spacer sequence that is complementary to crRNA. At this point, the complex will locate to the region of the PAM/ primary spacer sequence, and the DNA double strand will be untangled, forming an[6-8] R-Loop. The crRNA will hybridize with the complementary strand, while the other strand remains free. Subsequently, the Cas9 protein precisely cuts the third base in the 5 'terminal direction of PAM to form a flat terminal product, with the HNH domain of Cas9 responsible for cutting the complementary crRNA paired DNA strand and the RuvC domain responsible for cutting another non-complementary DNA strand. Finally, under the action of Cas9, DNA double strand breaks (DSB), and the expression of foreign DNA is silenced.
2.3 Deep learning and CRISPR-Cas9
Elektrum uses neural architecture search (NAS) and transfer learning techniques, combined with deep learning methods, to autonomously explore the architectural space of dynamically interpretable neural network (KINN) models, aiming to uncover the underlying dynamics of the system. The framework first generates a set of candidate KinNs through probabilistic modeling genetic algorithms to predict in vitro measurable motion velocities. The second NAS step then combines KINN with a convolutional neural network (CNN) to capture the series-dependent signals of subtle differences in the in-vivo data. The pre-trained KINN model acts as an intermediate layer to optimize the performance of the in vivo model. Elektrum's data-driven approach efficiently constructs multiple candidate dynamics models that go beyond human-derived models. Bayesian probabilistic genetic algorithm updates the model structure distribution according to [9]KINN performance, and shows advantages over random sampling in identifying accurate models of simulated dynamical systems. Using in vivo datasets as resistance, we evaluated the performance of a dynamic interpretable neural network (KINN) trained on in vitro data to predict the off-target dissociation rate of Cas9. The results show that the KINN model can accurately predict the dissociation rate, and there is a strong positive correlation between the predicted dissociation rate and the observed level of non-target editing. KINN performs better in vivo prediction than traditional Cas9 non-target prediction methods. The results of this study suggest that methods combined with AI deep learning can provide more accurate and reliable predictions of non-target Cas9 cleavage, providing important support for the safe use of Cas9 in research and therapeutic applications.
2.4 Application of CRISPR/Cas9 gene editing technology in medical field
CRISPR/Cas9 technology plays a crucial role in various aspects of disease research and treatment. Its integration with techniques like PCR and DNA sequencing enables rapid pathogen detection, facilitating timely disease management.
1) Application in infectious diseases: In the realm of infectious diseases, CRISPR/Cas9 has demonstrated significant potential. Specifically, it has been successfully employed to target the human immunodeficiency virus (HIV) genome, inhibiting viral replication, clearing infections, and even activating latent virus transcription to facilitate its elimination, offering promise for HIV treatment. CRISPR/Cas9 technology has expanded its applications to gene editing of various DNA viruses, including high-risk human papillomavirus [10](HPV), herpes virus, vaccinia virus, JC polyomavirus, avian adenovirus type 4, and African swine fever virus. 2) Application in tumor diseases: The CRISPR/Cas9 system is a breakthrough development in promoting tumor gene-level therapy, such as building tumor models, targeting the knockout of oncogene pathogenic sites, restoring the activity of tumor suppressor genes, reducing drug resistance of tumor cells, and conducting tumor immunotherapy. CRISPR/Cas9 technique mediated the gene knockout of peptidyl prolyl isomerase A[11](PPIA), which effectively increased the sensitivity of multiple myeloma cells to proteasome inhibitors. 3) Application in genetic diseases: The strategy of correcting point mutations associated with genetic diseases while preserving genetic function is one of the ideal ways to treat genetic diseases. In 2014, Yin et al. reported a study of CRISPR/ Cas9-mediated HDR correction of Fah gene point mutations in adult mice, demonstrating that correcting gene mutations using CRISPR/ Cas9-mediated gene editing is feasible in adult animals and has the potential to correct genetic mutations for human genetic diseases. This strategy has also been applied to the study of other similar genetic diseases. For example, Yang et al. successfully corrected the G→A point mutation of Otc gene exon in mice with the same technology, confirming the therapeutic effect of this therapy on neonatal hyperammonemia [12].
3 Application cases of deep learning in gene editing
3.1 Deep learning-based gene editing
Machine learning is to generalize and summarize existing data by generating models, thus mining the rules of existing data sets to achieve the purpose of predicting unknown data. An important direction of CRISPR/Cas9 research is the prediction of sgRNA targeting efficiency (Wang et al 2020). The application of machine learning in CRISPR/Cas system is mainly divided into 6 steps :1. Data acquisition, integrating SgRNA activity data of different experimental methods and different databases; 2. Data preprocessing and data normalization, eliminating redundant information and unreasonable data; 3. Feature engineering: Feature construction is carried out by pairing sgRNA sequence information with target sequence bases; 4. Feature selection, using feature selection algorithm to select features related to the target; 5. Train the model, calculate the evaluation scores of the training set and the verification set by cross-validation method to judge the model fitting and optimize the model parameter index; 6. Model evaluation: error analysis was carried out through relevant independent data to evaluate the accuracy and generalization ability of the model (Liu et al2020).
3.2 Attention mechanism analysis sgRNA sequence method
The analysis of sgRNA activity uses the spatiotemporal attention mechanism method, and the preferences for different sites of the target sequence are specifically demonstrated as follows:
For the target sequence X∈R, where l is the length of the target sequence, the following formula can be obtained for sgRNA activity y under the condition of satisfying linear regression:
\( y=AX \) (1)
Where 𝐴 ∈ 𝑅𝑑, then the derivative of the above formula is
\( dy=\sum_{i}^{d}A_{i}dX_{i} \) (2)
Where Ai, and Xi; Represents the i th dimension of the vector X and A, Ai represents the degree to which the function y changes with Xi at X, i.e. X; The importance of... Therefore, for Ai, the solution can be obtained by constructing a trainable non-negative weight vector x \( ∈R^{t} \) :
\( y=W×AX^{t} \) (3)
Where, the size of the preference coefficient matrix 𝐴𝑠 measures the importance of 𝑥𝑜ℎ and can be expressed as nucleotide preferences at different locations of sgRNA.
3.3 Predict the impact
In order to evaluate the effect of target flanker sequences on the prediction of Cas9 and its variant sgRNA activity, flanker sequences with lengths of 0, 1, 2, 3, 4 and 5nt on the left and right sides of target sites were taken (Figure 1A), and sequence fragments with lengths of 20, 22, 24, 26, 28 and 30 nt were obtained. Combined with three different sequence encoding methods, label encoding, one-hot encoding and two encoding, a total of 18 combination types are obtained (Figure 3-1B, C and D). For Cas9 and its variant sgRNA activity data sets, Sklearn toolkit was used to divide the sgRNA activity data sets according to the method of training set: test set =8:2, and the sequences in the training set and test set data were encoded according to 18 combination types to obtain 18 different training sets and test sets.
3.4 Data training
The sequence fragments in the training set are encoded as training data, and the activity of SgRNA is used as training labels. The multi-layer perceptron algorithm is used to learn the training data and training labels, and the learning parameters in the multi-layer perceptron algorithm are set as default parameters. The maximum iteration learning rounds are set to 5000 times. The multi-layer perceptron algorithm obtains 18 sets of models by learning 18 sets of different training data, and uses test sets to evaluate the prediction accuracy of sgRNA activity of the models. The specific method is to encode sequence fragments of the test set as test data, and the activity of SgRNA as test labels. The SgRNA prediction activity of the test set was obtained by using the model prediction test data as the prediction label, and the spearman coefficient between the prediction label and the test label was obtained.
3.5 Data results
In the SpCas9-VROR variant, when the sequence length is extended to 28nt, the prediction accuracy of sgRNA activity under ont-hot coding and two coding is higher than that of 20-26 nt sequence under the same coding. When the sequence length is extended to 30 nt, the prediction accuracy of SGRNA activity is higher than that of SGRNA activity under the same coding. The prediction accuracy of sgRNA activity under label coding, one-hot coding and two-coding is higher than that of other length sequences under the same coding (Figure 1).
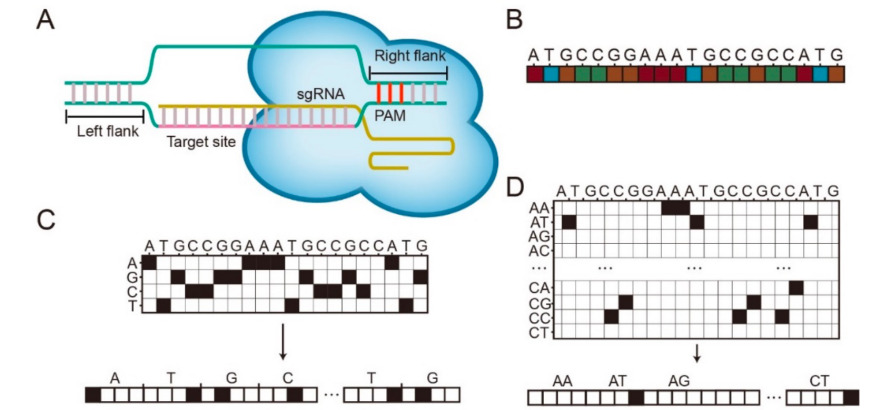
Figure 1. Evaluation of target flanker sequences against Cas9 results
(A) The position relationship between target flanker sequence and sgRNA sequence. The yellow fragment is the sgRNA sequence, the pink fragment is the target sequence, and the target sequence extending from left to right is the flanker sequence.
(B) Schematic diagram of label coding mode. The bases A, T, C and G in the sequence are coded respectively to correspond to four different colors.
(C) one-hot coding schematic, encoding the data of 1 × 4 bases in the sequence, for example, base A is encoded as [1,0,0,0], the black square in the figure represents the coded value of 1, and the white square represents the coded value of 0.
(D) Schematic diagram of two coding mode, encoding the two consecutive bases in the sequence into 1 × 16 data,
AT coding for,1,0,0,0,0,0,0,0,0,0,0,0,0,0,0 [0], for example, figure in the black squares encoding the value of 1, white squares said code value is 0.
In the xCas9 variant, when the sequence length was 28 nt and 30 nt, the prediction accuracy of sgRNA activity in the three coding cases exceeded that of other sequence lengths under the same coding. In the SpCas9-NG variant, when the sequence length was extended to 30 nt, the prediction accuracy of SgRNA activity under one-hot coding and two-coding was higher than that of 20-26 nt sequences under the same coding (Figure 3-1E). Comparatively, target flanker sequences had little effect on sgRNA prediction activity of Cas9 (Figure 3-1E). In conclusion, target flanker sequence can be used as one of the important features in predicting SgRNA activity of Cas9 variants, which helps to improve the accuracy of predicting sgRNA activity of some Cas9 variants.
3.6 Sequence and activity of Cas9 and its sgRNA variants
In order to analyze the location-dependent nucleotide preference of Cas9 and its variant SgRNA sequences for activity, an attention mechanism algorithm was introduced into the deep learning algorithm to evaluate the activity preference of the target SgRNA sequences at different positions by extracting matrix coefficients of the connection layer in the attention module. The steps of parsing SgRNA sequence based on attention mechanism algorithm are as follows :(1) The target sequence is encoded into a matrix with size of 21x4 through one-hot as input data; (2) Convolutional neural network was used to extract the eigenvalues of the input data, and the eigendata of different positions of the target sequence were extracted respectively through average pooling and maximum pooling methods; (3) Connect the characteristic data of the target sequence and standardize the data batch; (4) The standardized data is calculated by convolutional neural network as the matrix coefficient of the attention mechanism module; (5) Dot multiply the matrix coefficients with the input data matrix as the feature extraction vector of the target sequence; (6) The feature extraction vector is used as the feature information of the target sequence through the convolutional neural network layer, the flattening layer and the connecting layer; Finally, the SgRNA activity predicted score was output through three fully connected layers.
Spatial attention mechanism algorithm was used to analyze the location-dependent nucleotide preference of sgRNA sequences for sgRNA activity, mainly in analyzing the attention matrix in the model. The specific method was to use one-hot encoding of SgRNA sequences as training data for Cas9 and variant sgRNA activity datasets. sgRNA activity is used as a training label. The training data and training labels of Cas9 variants are learned by the algorithm to generate SgRNA activity prediction models for different Cas9 variants. The matrix attention coefficients of different positions are obtained from the models and averaged. The location-dependent nucleotide preferences of Cas9 and its variant sgRNA sequences for activity were obtained. In summary, deep learning technology provides new tools and methods for gene editing, which can improve editing efficiency, accuracy and repeatability. By combining machine learning and deep learning techniques, as well as attention mechanism analysis of sgRNA sequences, gene editing technology will be able to better meet future research and application needs.
4 Limitations and challenges of CRISPR/Cas9 gene editing
CRISPR/Cas9 technology as a revolutionary gene editing tool, although it brings great potential, but also faces a series of challenges. One of these is nonspecific splicing, in which the Cas9 protein is spliced outside the target site, leading to unexpected genomic changes. In addition, cytotoxicity is also an important issue, the use of CRISPR/Cas9 technology may lead to cell death or abnormal proliferation, affecting the experimental results and application effects.
4.1 The CRISPR/Cas9 technique is not specific cutting
Nonspecific cutting is a serious challenge in CRISPR/Cas9 technology and can lead to unpredictable genomic changes, which increases the risk and uncertainty of gene editing. Faced with this challenge, researchers are actively exploring the use of deep learning techniques to solve non-specific shear problems. [13] These models, based on extensive experimental data and genomic knowledge, are able to accurately predict the shear activity of CRISPR/Cas9 systems and help researchers select more reliable and safe editing targets. However, while deep learning models are excellent at predicting shear locations, there are still some challenges. One of them is the accuracy and generalization ability of the model. While models may perform well on specific data sets, they may perform differently under different conditions, especially when dealing with different kinds of cells or genomes. In summary, although deep learning technology provides a new way to solve the non-specific cutting problem in CRISPR/Cas9 technology, it still faces some challenges and limitations in practical applications.
4.2 Strategies for coping with cytotoxicity and other potential challenges
Cytotoxicity is another important challenge in CRISPR/Cas9 technology, with implications ranging from the effects of gene editing to the reliability of experimental results. During gene editing, overexpression or mistargeting of the Cas9 protein can lead to cell death or abnormal proliferation, seriously affecting the accuracy and repeatability of the experiment. The use of deep learning models by researchers to assess the effects of gene editing on tumor cells. By analyzing a large number of tumor cell biological characteristics and gene expression data, they built a predictive model that accurately predicted the effects of different editing regimens on tumor cell survival and proliferation. These models not only help researchers evaluate the safety of editing protocols, but also provide guidance for designing more efficient gene-editing experiments. Therefore, the use of deep learning techniques to predict the effects of gene editing on cell survival and proliferation is an important way to mitigate the cytotoxicity problems in CRISPR/Cas9 technology. The establishment of these models can not only improve the safety and efficiency of gene editing experiments, but also provide important reference and guidance for research in related fields.
5 Conclusion
CRISPR technology has broad application prospects in disease treatment, gene function regulation, drug development and other aspects, but there are still some problems that limit its application in the actual environment. For example, off-target effects, safety problems caused by autoimmunity, and limitations of [14]PAM sequences. However, with the development of recent years, CRISPR/Cas9 technology has been continuously improved, compared with the previous, both editing efficiency and avoiding off-target effects have been significantly improved, especially in clinical application, which is expected to solve some diseases that have no treatment drugs before or some genetic defects. The rapid development of gene editing technology has brought great breakthroughs and possibilities to the field of life science, especially CRISPR/Cas9 technology as a revolutionary gene editing tool. However, with the application of the technology, CRISPR/Cas9 technology also faces a series of challenges, such as non-specific cutting and cytotoxicity issues. Although deep learning technology provides new ideas and methods to solve these challenges, there are still challenges in model accuracy and generalization ability, which need further research and improvement. Therefore, future research should focus on improving the accuracy and interpretability of models, developing more effective training methods and data acquisition strategies to better address challenges in gene editing, and driving the development and application of the field.
References
[1]. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. https://doi.org/10.1126/science.1225829
[2]. K. Xu, X. Wang, Z. Hu and Z. Zhang, "3D Face Recognition Based on Twin Neural Network Combining Deep Map and Texture," 2019 IEEE 19th International Conference on Communication Technology (ICCT), Xi'an, China, 2019, pp. 1665-1668, doi: 10.1109/ICCT46805.2019.8947113.
[3]. Nobel Prize 2020 in Chemistry honors CRISPR: a tool for rewriting the code of life.
[4]. Zheng, Jiajian, et al. "The Random Forest Model for Analyzing and Forecasting the US Stock Market in the Context of Smart Finance." arXiv preprint arXiv:2402.17194 (2024).
[5]. Yang, Le, et al. "AI-Driven Anonymization: Protecting Personal Data Privacy While Leveraging Machine Learning." arXiv preprint arXiv:2402.17191 (2024).
[6]. Yang Yang, WANG Fenglin, Liu De et al. Research progress of CRISPR⁃Cas9 technology in Production of plant secondary Metabolites [J]. Advances in Biotechnology, 2002,12(06):806-816.
[7]. Jiang, Fuguo, and Jennifer A. Doudna. "CRISPR–Cas9 structures and mechanisms." Annual review of biophysics 46 (2017): 505-529.
[8]. Redman, Melody, et al. "What is CRISPR/Cas9?." Archives of Disease in Childhood-Education and Practice 101.4 (2016): 213-215.
[9]. Zhu, Mengran, et al. "Utilizing GANs for Fraud Detection: Model Training with Synthetic Transaction Data." arXiv preprint arXiv:2402.09830 (2024).
[10]. Wu, Jiang, et al. "Data Pipeline Training: Integrating AutoML to Optimize the Data Flow of Machine Learning Models." arXiv preprint arXiv:2402.12916 (2024).
[11]. Yu, Hanyi, et al. "Machine Learning-Based Vehicle Intention Trajectory Recognition and Prediction for Autonomous Driving." arXiv preprint arXiv:2402.16036 (2024).
[12]. Huo, Shuning, et al. "Deep Learning Approaches for Improving Question Answering Systems in Hepatocellular Carcinoma Research." arXiv preprint arXiv:2402.16038 (2024).
[13]. Ma, Yuanwu, Lianfeng Zhang, and Xingxu Huang. "Genome modification by CRISPR/Cas9." The FEBS journal 281.23 (2014): 5186-5193.
[14]. Zhang, Jian-Hua, et al. "Optimization of genome editing through CRISPR-Cas9 engineering." Bioengineered 7.3 (2016): 166-174.
Cite this article
Cui,Z.;Lin,L.;Zong,Y.;Chen,Y.;Wang,S. (2024). Precision gene editing using deep learning: A case study of the CRISPR-Cas9 editor. Applied and Computational Engineering,64,133-140.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 6th International Conference on Computing and Data Science
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. https://doi.org/10.1126/science.1225829
[2]. K. Xu, X. Wang, Z. Hu and Z. Zhang, "3D Face Recognition Based on Twin Neural Network Combining Deep Map and Texture," 2019 IEEE 19th International Conference on Communication Technology (ICCT), Xi'an, China, 2019, pp. 1665-1668, doi: 10.1109/ICCT46805.2019.8947113.
[3]. Nobel Prize 2020 in Chemistry honors CRISPR: a tool for rewriting the code of life.
[4]. Zheng, Jiajian, et al. "The Random Forest Model for Analyzing and Forecasting the US Stock Market in the Context of Smart Finance." arXiv preprint arXiv:2402.17194 (2024).
[5]. Yang, Le, et al. "AI-Driven Anonymization: Protecting Personal Data Privacy While Leveraging Machine Learning." arXiv preprint arXiv:2402.17191 (2024).
[6]. Yang Yang, WANG Fenglin, Liu De et al. Research progress of CRISPR⁃Cas9 technology in Production of plant secondary Metabolites [J]. Advances in Biotechnology, 2002,12(06):806-816.
[7]. Jiang, Fuguo, and Jennifer A. Doudna. "CRISPR–Cas9 structures and mechanisms." Annual review of biophysics 46 (2017): 505-529.
[8]. Redman, Melody, et al. "What is CRISPR/Cas9?." Archives of Disease in Childhood-Education and Practice 101.4 (2016): 213-215.
[9]. Zhu, Mengran, et al. "Utilizing GANs for Fraud Detection: Model Training with Synthetic Transaction Data." arXiv preprint arXiv:2402.09830 (2024).
[10]. Wu, Jiang, et al. "Data Pipeline Training: Integrating AutoML to Optimize the Data Flow of Machine Learning Models." arXiv preprint arXiv:2402.12916 (2024).
[11]. Yu, Hanyi, et al. "Machine Learning-Based Vehicle Intention Trajectory Recognition and Prediction for Autonomous Driving." arXiv preprint arXiv:2402.16036 (2024).
[12]. Huo, Shuning, et al. "Deep Learning Approaches for Improving Question Answering Systems in Hepatocellular Carcinoma Research." arXiv preprint arXiv:2402.16038 (2024).
[13]. Ma, Yuanwu, Lianfeng Zhang, and Xingxu Huang. "Genome modification by CRISPR/Cas9." The FEBS journal 281.23 (2014): 5186-5193.
[14]. Zhang, Jian-Hua, et al. "Optimization of genome editing through CRISPR-Cas9 engineering." Bioengineered 7.3 (2016): 166-174.









