1. Introduction
Medical image segmentation [1, 2] is a critical research problem in the fields of computer vision and medical image processing. However, medical image segmentation faces numerous challenges, particularly in dealing with issues like noise, low contrast, and the complexity of target structures, all of which can negatively impact the accuracy of segmentation results.
In recent years, deep learning has made significant advancements in medical image segmentation tasks. U-Net’s [3] impressive performance on many problems, however, there is still room for improvement when dealing with challenging medical image tasks, and the acquisition and processing of medical images may introduce various biases that need to be properly addressed during the segmentation process. Specifically, U-Net may not fully capture complex contextual information and long-range dependencies in the images, leading to reduced segmentation accuracy.
To address these issues, we propose model called GSANet. This model adopts the U-shaped architecture and introduces several innovative modules to overcome the limitations of U-Net. These new modules include the Graph Self-Attention (GSA) module and the Global Interaction Block. The GSA module effectively captures long-range dependencies and complex contextual information in the images, while the Global Interaction Block enhances the model's representational capacity by complementing and integrating features. Moreover, the attention block improves segmentation accuracy by allowing the model to focus more on key information in the images.
We evaluate the GSANet model on the DRIVE dataset and ISIC2018 dataset. Our experimental results demonstrate that by using GSANet, our model achieves significant performance improvements on these tasks.
The primary contributions of this paper are outlined as follows:
1. Our model leverages the strengths of U-Net while introducing novel modules, including the Graph Self-Attention (GSA) module, Cross Update Block, and Attention Block. These modules address the limitations of U-Net in handling challenging medical image tasks.
2. The experimental results demonstrate significant performance improvements achieved by our model on these challenging medical image segmentation tasks. These findings validate the effectiveness of our designed modules and highlight the superiority of our model.
In the subsequent sections, we provide an elaborate description of our proposed deep learning model and its innovative modules. We then present the experimental setup, results, and discussions, followed by conclusions and future research.
2. Related work
2.1. Unet
U-Net has been widely used for various image segmentation tasks since its introduction in 2015.The key innovations of U-Net lie in its symmetric encoder-decoder architecture and skip connections. The symmetric encoder-decoder architecture allows the network to learn features at different resolution levels, while skip connections enable the network to reuse features from low-resolution levels at higher-resolution levels. These two design aspects together allow the network to retain fine-grained details of the images while also capturing contextual information.
While U-Net has exhibited strong performance across numerous medical image segmentation tasks, there remains potential for enhancement in addressing challenging medical image tasks. For instance, U-Net may not provide satisfactory results when dealing with medical images that contain complex contextual information and long-range dependencies. This is mainly because the skip connections in U-Net may not effectively capture long-range contextual dependencies, and its encoder-decoder architecture may encounter difficulties in capturing complex contextual information. Therefore, improving U-Net to better address these challenging tasks remains an important research question in the field of medical image segmentation.
2.2. Graph Convolutional Network (GCN)
Graph Convolutional Network (GCN) [4] is a neural network tailored for graph data. GCN's core concept revolves around efficiently integrating neighboring information of nodes within the graph via graph convolutional operations, thereby capturing intricate dependencies inherent in the graph structure. In this process, graph nodes are regarded as carriers of signals, and graph edges represent the paths for signal propagation. GCN achieves local signal propagation and integration on the graph by defining graph convolutional operations. In the original GCN model, graph convolutional operations are defined as a linear combination of node features and their neighboring node features. Specifically, given a graph \( G=(V,E) \) , where \( V \) is the set of nodes and \( E \) is the set of edges,for each node \( {v_{i}}∈V \) , its feature \( {h_{i}} \) is updated as the weighted average of its neighboring node features:
\( h_{i}^{ \prime }=\sum _{j∈N(i)}{w_{ij}}{h_{j}} \) (1)
where \( {N_{(i)}} \) is the set of neighboring nodes of node \( {v_{i}} \) ,and \( {w_{ij}} \) is the weight of the edge between node \( {v_{i}} \) and node \( {v_{j}} \) which controls the influence of different neighboring nodes on the current node.
However, traditional GCN may struggle to handle medical images with complex feature distributions and noisy interference. This is because traditional GCN only considers first-order neighbor information of the graph, neglecting higher-order neighbor information. Additionally, traditional GCN does not take into account node-specific information, which could lead to information loss. Moreover, when the graph structure is very complex or the graph scale is large, the effectiveness of GCN may be compromised. Therefore, improving GCN to better address these issues is also an important research question in the current context.
3. Method
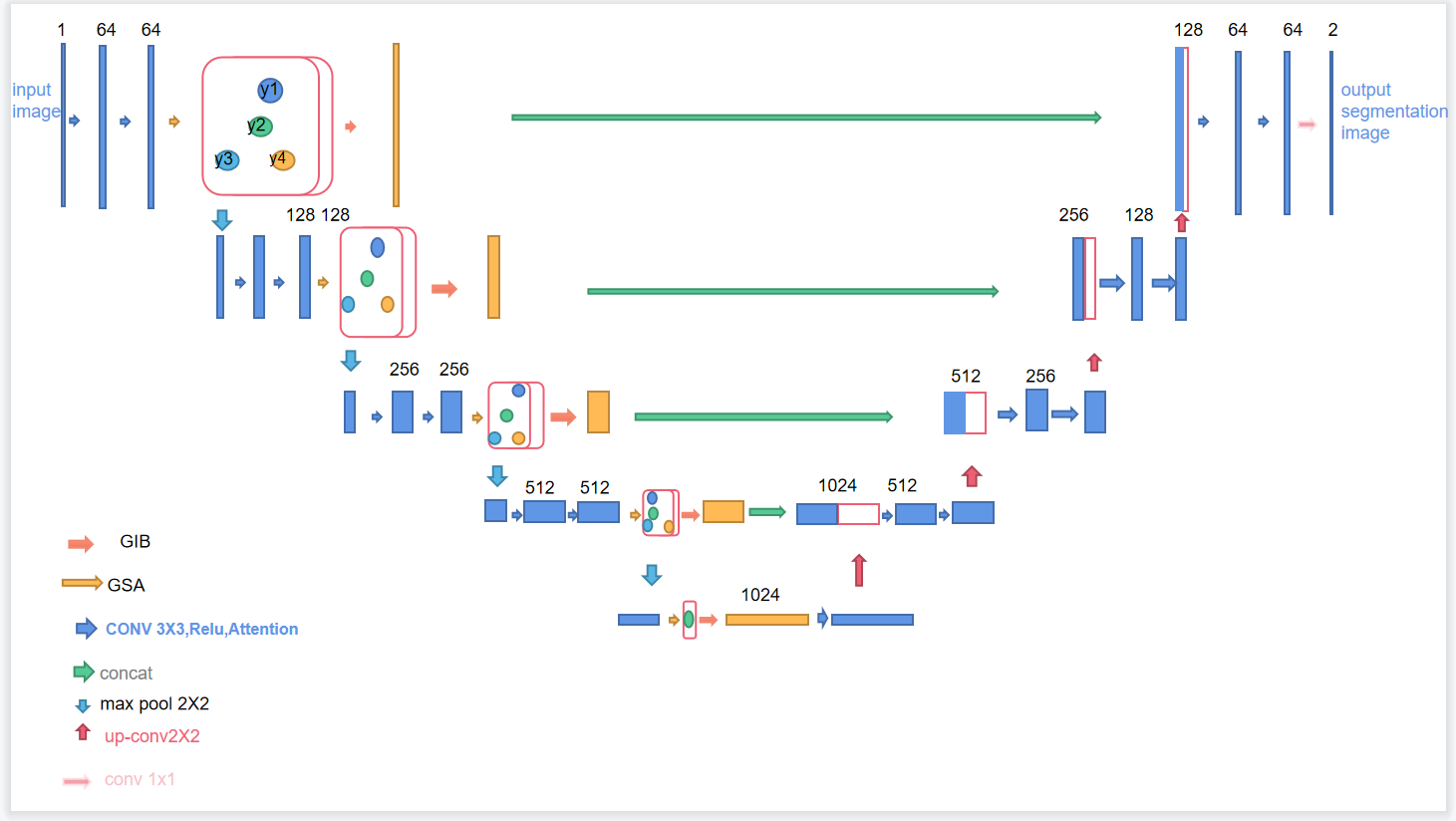
Figure 1. A brief description of our proposed model
As shown in Fig 1, our GSANet model is based on the U-Net architecture and introduces two innovative modules: Graph Self-Attention (GSA) and Global Interaction Block (GIB). These modules are integrated into the U-Net encoder to address the limitations of U-Net in handling complex image contexts and long-range dependencies. The GSA module captures long-range dependencies and complex contextual information in images, while the GIB module enhances the model's representation capabilities by integrating global, local, high-level, and low-level features. This design aims to improve the model's understanding of internal image structures and its robustness to input biases, optimizing performance in challenging tasks such as medical image segmentation.
3.1. Graph Self-Attention
For the first time, we introduce the Graph Self-Attention (GSA) module, as shown in the Fig 2 which is based on the self-attention mechanism [5] of Graph Convolutional Networks (GCN), to effectively capture long-range dependencies and complex contextual information in the image. In the GSA module, we first calculate a similarity matrix based on the input feature map, which measures the similarity between each pair of pixels. The computation of this similarity matrix can be described using the following equation:
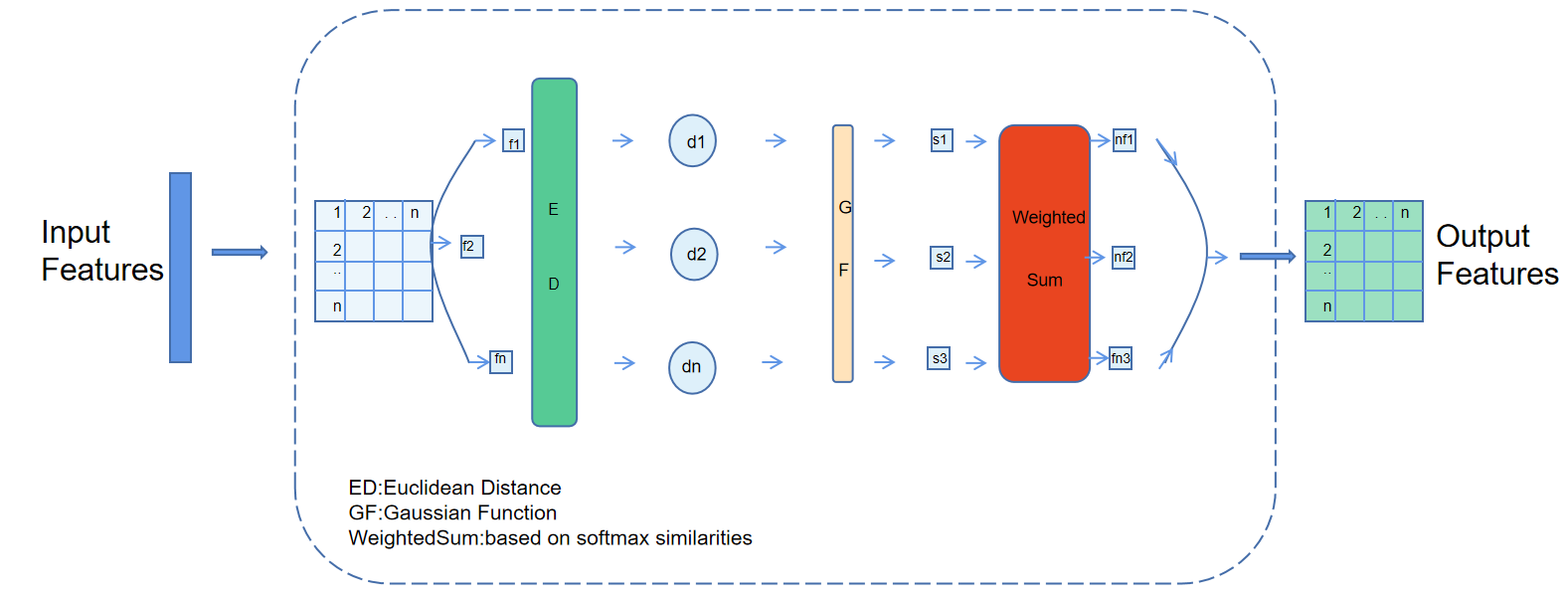
Figure 2. A brief description of Graph Self-Attention block
\( S=\frac{{e^{ \prime (-\frac{||{h_{i}}-{h_{j}}||_{2}^{2}}{{σ^{2}}})}}}{\sum _{k=1}^{N}{e^{(-\frac{||{h_{i}}-{h_{j}}||_{2}^{2}}{{σ^{2}}})}}} \) (2)
This formula essentially uses a Gaussian kernel function [6] to compute the similarity between each pair of pixels and normalizes it. Subsequently, this similarity matrix is used to update the feature map. The new feature of each pixel is a weighted sum of its neighboring features, where the weights are determined by the similarity matrix. This updating process can be described by the following formula:
\( h_{i}^{ \prime }=\sum _{j=1}^{N}{S_{ij}}{h_{j}} \) (3)
This formula effectively performs a weighted sum of features, where the weights are determined by the similarity matrix. This mechanism allows each pixel to receive information from the entire image, efectively capturing long-range dependencies and complex contextual information.
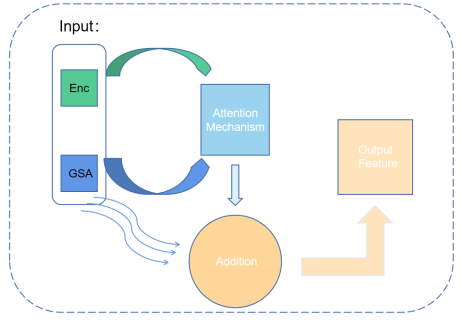
Figure 3. A brief description of Global Interaction Block
3.2. Global Interaction Block
In our research, we found that the simple attention mechanism did not fully utilize the global and local information in the image, as well as the high-level and low-level features. Therefore, we proposed a new module called Global Interaction Block (GIB). This module combines the attention mechanism and cross-update strategy, which can use global and local information, as well as high-level and low-level features at each level. The design philosophy of the GIB originates from the concept of inductive bias. In the GIB, our inductive bias is to let the model learn and use global and local features, as well as high-level and low-level features simultaneously. This design enables the model to learn and predict on multiple scales, thereby improving the model’s generalization ability and performance. In the GIB, we first process the feature image through the self-attention mechanism to extract global and local feature information. Then, we use a cross-update strategy to fuse these feature information together. Specifically, we use a soft attention mechanism to average the global features and local features by weight, generating a new feature map. Finally, this new feature map is fused with the original feature map to generate the final output feature map. Let there be feature maps G and E, both with dimensions of C × H × W , where C represents the number of channels, and H and W are the height and width of the feature maps, respectively. First, we flatten the feature maps G and E to C ×N, where N = H ×W, resulting in Gflat and Eflat. Subsequently, we compute the matrix multiplication of Eflat and Gflat and perform softmax normalization along the last dimension to obtain the class-specific feature Fc:
\( {F_{c}}=softmax{(}\frac{E_{flat}^{T}\cdot {G_{flat}}}{\sqrt[]{C}}) \) (4)
Following this, we perform matrix multiplication of Fc and Eflat , and reshape it back to the original spatial dimensions C×H×W , resulting in the class-specific feature F :
\( F=Reshape({F_{c}}\cdot E_{flat}^{T},(C,H,W)) \) (5)
Here, the reshaping operation is defined as follows:
\( {F_{ijk}}={F_{fla{t_{jk}}}},i∈\lbrace 1,2,....,C\rbrace ,j,k∈\lbrace 1,2,....,N\rbrace \) (6)
Finally, we perform element-wise addition of the class-specific feature \( F \) and the original feature map \( G \) to produce the final output feature map \( O \) :
\( O=F+G \) (7)
4. Experiments
4.1. Experimental Dataset and Parameter Settings
The experiments utilized two medical image datasets, DRIVE [7] and ISIC2018 [8, 9], for retinal and skin pathology image segmentation tasks, respectively. These datasets contain complex medical images with unclear and blurry edges. To efficiently leverage limited data, we divided them into training and validation sets in a 9:1 ratio. Training and validation were conducted using the PyTorch framework, employing the SGD optimization [10] algorithm with an initial learning rate of 0.01. During training, the model was guided by the kl loss function [11].
Table 1. Model Evaluations
Model | Dataset | DICE | Miou |
Unet[3] | DRIVE | 81.31 | 81.60 |
ISIC18 | 72.14 | 74.55 | |
Unet++[12] | DRIVE | 82.44 | 82.63 |
ISIC18 | 78.32 | 75.12 | |
g-U-Nets[13] | DRIVE | 83.17 | 84.21 |
ISIC18 | 78.94 | 79.13 | |
TransUnet[14] | DRIVE | 84.16 | 85.35 |
ISIC18 | 81.66 | 83.51 | |
GSANet | DRIVE ISIC18 | 86.13 83.74 | 87.56 84.18 |
4.2. Experimental Results
Table 1 shows the performance of our GSANet model and other comparative models in terms of Dice coefficient and Miou on the DRIVE and ISIC2018 datasets. The results indicate that our GSANet model outperforms all other models, achieving the highest Dice coefficient and Miou on both the DRIVE and ISIC2018 datasets. This demonstrates that our model accurately identifies and segments the vascular structures in retinal images and effectively recognizes the shapes of skin lesions, even when the boundaries between the lesion regions and the surrounding healthy skin are not well-defined.
Through comparisons with other models, we observed that the GSANet model exhibits more significant advantages when dealing with complex images. This can be attributed to the effectiveness of the GSA and GIB modules in our model, which can extract and integrate both local fine details and global contextual information from the images, resulting in more accurate segmentation.
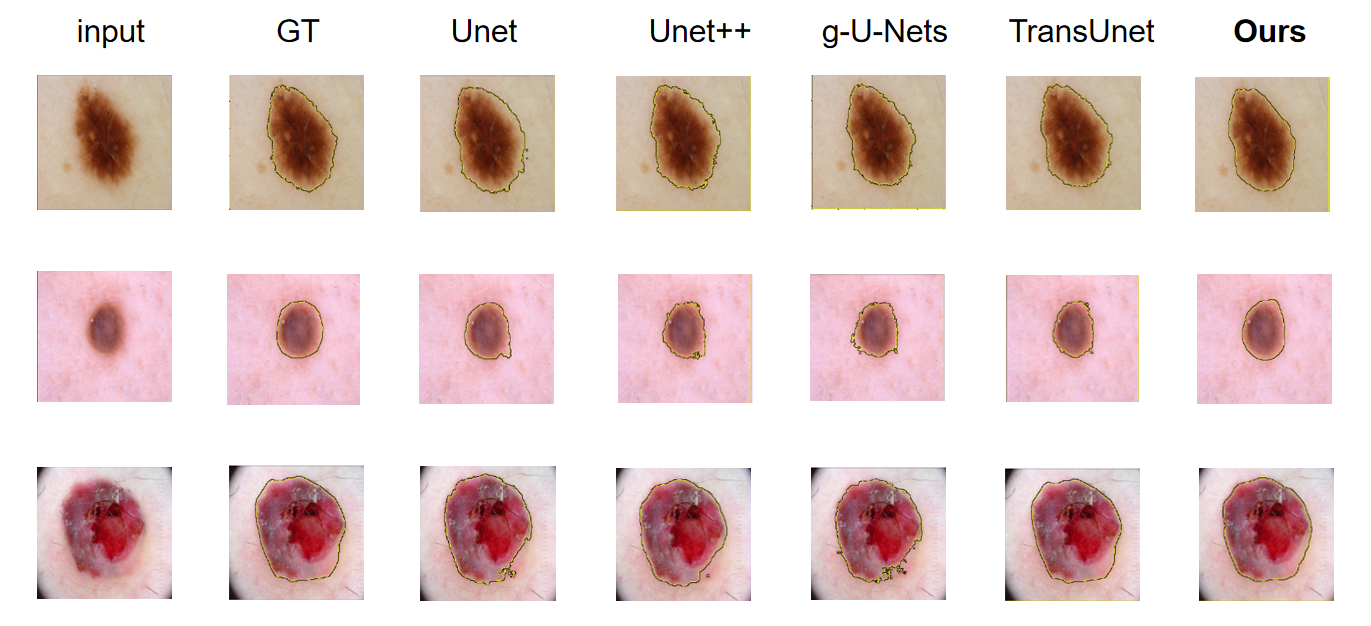
Figure 4. comparison of different approaches by visualization.
Moreover,from the visualization results in Fig 4, we can observe that the segmentation results of the GSANet model are very close to the Ground Truth. Our model accurately identifies and segments the target structures, even in cases where the boundaries of the target structures are unclear or mixed with background information. Even when other models struggle to segment the detailed parts, the GSANet model can still accurately delineate the boundaries of the target. By fully leveraging the global and local information of the images, as well as high and low-level features, this once again demonstrates the outstanding performance of our model in handling complex medical image data.
5. Conclusion
In this study, we present a new model GSANet, dedicated to addressing the challenges in medical image segmentation. GSANet integrates the U-Net architecture with the innovative Graph Self-Attention (GSA) module and Global Interaction Block (GIB), demonstrating significant superiority over existing methods in handling complex medical tasks such as retinal and skin pathology image segmentation. The GSA module effectively captures long-range dependencies and complex contextual information in images, providing crucial support for understanding the structure and texture of medical images. Simultaneously, the GIB module enhances the model's understanding by integrating global and local feature information. While GSANet excels in medical image processing, there is still room for improvement. Currently, the model focuses on global and local information, and future research can delve deeper into exploring high-level and low-level features to further enhance performance.
References
[1]. Chen, C.T., C.K. Tsao, and W.C. Lin, Medical image segmentation by a constraint satisfaction neural network. IEEE Transactions on Nuclear Science, 2016. 38(2): p. 678-686.
[2]. Jian, M., et al., Dual-Branch-UNet: A Dual-Branch Convolutional Neural Network for Medical Image Segmentation. Computer Modeling in Engineering and Science, 2023.
[3]. Ronneberger, O., P. Fischer, and T. Brox. U-Net: Convolutional Networks for Biomedical Image Segmentation. in Springer, Cham. 2015.
[4]. Lei, F., et al., Graph convolutional networks with higher‐order pooling for semisupervised node classification. Concurrency and Computation: Practice and Experience, 2020.
[5]. Vaswani, A., et al., Attention Is All You Need. arXiv, 2017.
[6]. Rasmussen, C.E. and C.K.I. Williams, Gaussian Processes for Machine Learning. 2005: Gaussian Processes for Machine Learning.
[7]. Araújo, R.J., J.S. Cardoso, and H.P. Oliveira. A deep learning design for improving topology coherence in blood vessel segmentation. in Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, October 13–17, 2019, Proceedings, Part I 22. 2019. Springer.
[8]. Codella, N., et al., Skin lesion analysis toward melanoma detection 2018: A challenge hosted by the international skin imaging collaboration (isic). arXiv preprint arXiv:1902.03368, 2019.
[9]. Tschandl, P., C. Rosendahl, and H. Kittler, The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Scientific data, 2018. 5(1): p. 1-9.
[10]. eon Bottou, L., Online learning and stochastic approximations. Online learning in neural networks, 1998. 17(9): p. 142.
[11]. He, Y., et al. Bounding box regression with uncertainty for accurate object detection. in Proceedings of the ieee/cvf conference on computer vision and pattern recognition. 2019.
[12]. Zhou, Z., et al. Unet++: A nested u-net architecture for medical image segmentation. in Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 20, 2018, Proceedings 4. 2018. Springer.
[13]. Gao, H. and S. Ji. Graph u-nets. in international conference on machine learning. 2019. PMLR.
[14]. Chen, J., et al., Transunet: Transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:2102.04306, 2021.
Cite this article
Wang,Z.;Liu,S.;Qin,C.;Duan,Z.;Ge,X.;Sun,F. (2024). GSANet: Enhancing accuracy in the segmentation of challenging medical images. Applied and Computational Engineering,75,66-72.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 2nd International Conference on Software Engineering and Machine Learning
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Chen, C.T., C.K. Tsao, and W.C. Lin, Medical image segmentation by a constraint satisfaction neural network. IEEE Transactions on Nuclear Science, 2016. 38(2): p. 678-686.
[2]. Jian, M., et al., Dual-Branch-UNet: A Dual-Branch Convolutional Neural Network for Medical Image Segmentation. Computer Modeling in Engineering and Science, 2023.
[3]. Ronneberger, O., P. Fischer, and T. Brox. U-Net: Convolutional Networks for Biomedical Image Segmentation. in Springer, Cham. 2015.
[4]. Lei, F., et al., Graph convolutional networks with higher‐order pooling for semisupervised node classification. Concurrency and Computation: Practice and Experience, 2020.
[5]. Vaswani, A., et al., Attention Is All You Need. arXiv, 2017.
[6]. Rasmussen, C.E. and C.K.I. Williams, Gaussian Processes for Machine Learning. 2005: Gaussian Processes for Machine Learning.
[7]. Araújo, R.J., J.S. Cardoso, and H.P. Oliveira. A deep learning design for improving topology coherence in blood vessel segmentation. in Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, October 13–17, 2019, Proceedings, Part I 22. 2019. Springer.
[8]. Codella, N., et al., Skin lesion analysis toward melanoma detection 2018: A challenge hosted by the international skin imaging collaboration (isic). arXiv preprint arXiv:1902.03368, 2019.
[9]. Tschandl, P., C. Rosendahl, and H. Kittler, The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Scientific data, 2018. 5(1): p. 1-9.
[10]. eon Bottou, L., Online learning and stochastic approximations. Online learning in neural networks, 1998. 17(9): p. 142.
[11]. He, Y., et al. Bounding box regression with uncertainty for accurate object detection. in Proceedings of the ieee/cvf conference on computer vision and pattern recognition. 2019.
[12]. Zhou, Z., et al. Unet++: A nested u-net architecture for medical image segmentation. in Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 20, 2018, Proceedings 4. 2018. Springer.
[13]. Gao, H. and S. Ji. Graph u-nets. in international conference on machine learning. 2019. PMLR.
[14]. Chen, J., et al., Transunet: Transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:2102.04306, 2021.









