1. Introduction
In the world today, lung cancer is still among the most prevalent cancer kinds. For this reason, in the medical world, early lung cancer detection and treatment are crucial. In today's developing era of artificial intelligence, it is possible to attempt an initial assessment of whether a patient has lung cancer based on different characteristics exhibited by the patient through models [1]. This not only allows patients to perform simple self-checks through the release of the model but also provides the medical community with a large amount of data of research value.
As artificial intelligence continues to develop, artificial intelligence (AI) has also begun to be applied [2]. Convolutional and recurrent neural networks, when paired with AI-based techniques, may be particularly useful in 2022 for increasing the precision of lung cancer prediction, as Chen et al [3]. In 2020, Alexander and others utilized an artificial intelligence matching system that could screen cancer patients efficiently and accurately [4]. Wu and colleagues, meantime, demonstrated how an AI-assisted system can be a useful and successful method to get around the difficulties associated with the Programmed Death-Ligand 1 evaluation [5]. Utilizing 3D deep learning techniques to process computed tomography (CT) images, Li and his team carried out a retrospective study in China in 2019.Their AI system achieved 75% sensitivity, 82% specificity. This study provides more precise and unbiased results and reducing the interpretation time of radiologists in the diagnosis of pulmonary nodules [6]. In 2018, researchers from the United States, led by Choi, conducted a retrospective study using Support Vector Machines. These results demonstrate the potential of AI in significantly improving the accuracy of lung cancer detection [7]. The research of Baldwin and his team shows the Local Context Pooling - Convolutional Neural Network is better than Brock model [8]. Overall, these findings highlight the potential of AI in improving the accuracy and efficiency of lung cancer diagnosis, which will ultimately benefit both patients and healthcare professionals significantly.
Improve lung cancer patient screening by developing bidirectional Long Short-Term Memory (LSTM) and bidirectional Gated Recurrent Unit (GRU) models using the Lung Cancer dataset from Kaggle is the capital aim with extraordinary research value. This dataset encompasses 16 features related to patient symptoms and lung cancer status, offering a diverse range of symptoms to enhance model accuracy. The approach advances AI-driven healthcare by integrating sophisticated machine learning techniques to refine diagnostic capabilities [9].
The methodology involves four key steps: preprocessing the dataset for model compatibility, defining the model architecture with bidirectional LSTM and GRU layers, training the model for 50 epochs, and evaluating the model's performance. The results indicate an accuracy of 52.17%, with accuracy, recall, and F1 scores for both cancerous and non-cancerous categories approximating 50%. While the hybrid model demonstrates average performance, it provides a foundation for further refinement. Optimizing model parameters and exploring additional techniques to enhance prediction accuracy and clinical relevance should be done in the future.
2. Methodology
2.1. Dataset description and preprocessing
This dataset is sourced from Kaggle [10]. The dataset illustrates the diversity of lung cancer symptoms. It mainly includes three categories: patient demographic information, medical records, and clinical data. These three categories encompass a total of 16 features and lung cancer. Before training the model, it is necessary to preprocess the experimental data. Smoking is a major and nonnegligible risk factor for lung cancer, and the longer the smoking duration and the greater the amount of smoking, the higher the risk of lung cancer. Therefore, the feature of age multiplied by smoking is used to obtain the smoking duration as the 17th feature. Additionally, noise is added to the dataset and standardization is performed. To make the data conform to the input requirements of the LSTM model, it is also necessary to transform the data from two dimensions to three dimensions.
2.2. Proposed approach
The core of this study mainly focuses on combining traditional logistic regression problems with models capable of processing time series data, such as LSTM and GRU. This integration enables the model to extract context and better handle classification issues. The specific process shown is analyzed in Figure 1.
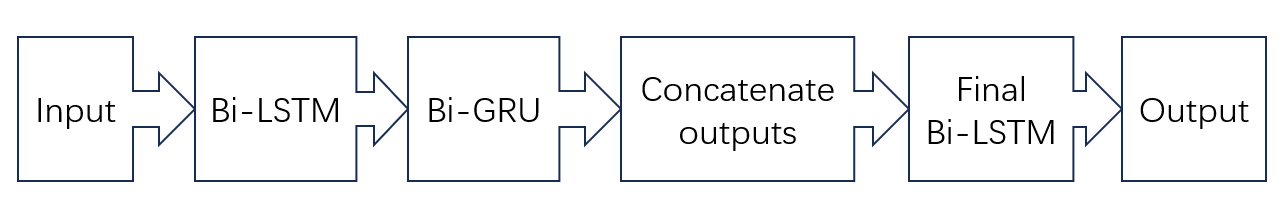
Figure 1. The pipeline of the model.
2.2.1. Bi-LSTM. The LSTM is an improved network structure that combines forward and backward information at each time step. Traditional LSTM networks can only process input sequences starting from the beginning and propagate information step by step. This means that when processing time series data, the network can only utilize information from earlier steps and cannot leverage subsequent information. To address this issue, the bidirectional LSTM introduces another LSTM network that processes the sequence in reverse. This allows for the utilization of both forward and backward information at every time step. Specifically, the bidirectional LSTM consists of two LSTM networks: one responsible for processing forward information and the other for processing backward information. The outputs of these two networks are merged at each time step, forming a global state that incorporates both forward and backward information. This structure enables the bidirectional LSTM to deal with sequences such as text, speech, and video, where the bidirectional LSTM exhibits improved performance.
2.2.2. Bi-GRU. The GRU is a variant of the Recurrent Neural Network (RNN), similar to the bidirectional LSTM. The bidirectional GRU is capable of considering as well as dealing with the forward and the backward information input sequence simultaneously, allowing it to capture a more comprehensive context when processing time series data. The structure of the bidirectional GRU includes two GRU networks, one responsible for processing forward information and the other for processing backward information. The outputs of these two GRU networks are merged at each time step, forming a global state that incorporates both forward and backward information.
2.2.3. Loss function. The binary cross-entropy loss function is suitable for situations where the output results of model are probability value, which is particularly common in binary classification problems. When the output layers use the Sigmoid activation function, the output values range between 0 and 1, which corresponds exactly to the probability value of the true label. At the same time, the value of the binary cross-entropy loss function is always non-negative. The binary cross-entropy loss function has good mathematical properties, making it easy to calculate and optimize:
 (1)
(1)
In the formula above, represents the true class of the i-th sample, which is typically 0 (negative class) or 1 (positive class). For each sample, the loss function calculates the cross-entropy between the true label and the predicted probability. If the true label is 1, then the loss is \( -log{(}{\hat{y}_{i}}) \) ; if the true label is 0, then the loss is \( -log{(}1-{\hat{y}_{i}}) \) :
2.3. Implementation details
In the model hyperparameter selection of this experiment, the batch size is set and gradually adjusted to 32, and the epochs is set to 50. To reduce the impact of overfitting, a Dropout layer was added to each layer, and the dropout rate for each Dropout layer was set to 0.2. This model uses the Adam optimizer This allows Adam to adjust as well as judge the learning speed of each parameter.
3. Result and Discussion
In this experiment, a hybrid model consisting of LSTM and GRU was applied to a test set with 600 samples, each equipped with complete labels. The accuracy evaluation results of the hybrid model will be provided in Table 1.
Table 1. Evaluate result.
precision | recall | f1-score | support | |||
0 | 0.52 | 0.55 | 0.54 | 296 | ||
1 | 0.54 | 0.50 | 0.52 | 304 | ||
accuracy | 0.53 | 600 | ||||
macro avg | 0.53 | 0.53 | 0.53 | 600 | ||
Weighted avg | 0.53 | 0.53 | 0.53 | 600 | ||
Table 1 shows the comprehensive evaluation results of the model in this experiment. The accuracy rate of the hybrid model reached 52.67%, which means the model accurately predicted 316 out of 600 samples. At the same time, this experiment used three performance metrics to evaluate the prediction effect on the positive and negative classes. Among the three-evaluation metrics, precision represents the proportion of samples that are actually class 0 (1) among all samples predicted as class 0 (1) by model. Recall indicates the proportion of samples correctly predicted as class 0 (1) by model.
The f1-score is providing a comprehensive and improvable indicator of precision and recall. Support represents the number of samples of class 0 (1) in the test set. Macro Average is a method for calculating the performance metrics of a classification model, which obtains an overall performance metric by calculating the arithmetic mean of the performance metrics for all classes. In this model evaluation, the values of the three-evaluation metrics are all 0.53. The Weighted Average metric is a method that considers class imbalance when evaluating. It calculates the average by assigning a weight to the performance metrics of each class, which is usually the relative frequency of the class in the dataset. Overall, the model in this experiment is at a moderate level, with a not-so-high prediction accuracy, and there are many aspects that can be optimized.
4. Conclusion
The primary contribution of this study is the integration of traditional logistic regression problems with advanced models like LSTM and GRU. However, the results reveal that these time-series models, while adept at capturing long-term dependencies, do not perform optimally on logistic regression tasks where feature relationships are less dynamic. The LSTM and GRU models struggled because they are designed for sequential data with contextual relationships, whereas the dataset in this study lacked such context, limiting the models' effectiveness. Traditional machine learning methods proved more suitable for this type of problem. Future research should investigate the synergy between logistic regression and sequential models, focusing on optimizing these models to better address logistic regression challenges. Additionally, exploring advanced deep learning architectures could offer solutions for more complex issues, enhancing the overall capability of predictive models.
References
[1]. Chiu H Y Chao H S Chen Y M 2022 Application of artificial intelligence in lung cancer Cancers vol 14 no 6 p 1370
[2]. Kanan M Alharbi H Alotaibi N et al. 2024 AI-Driven Models for Diagnosing and Predicting Outcomes in Lung Cancer: A Systematic Review and Meta-Analysis Cancers vol 16 no 3 pp 674
[3]. Chen S 2022 Models of Artificial Intelligence‐Assisted Diagnosis of Lung Cancer Pathology Based on Deep Learning Algorithms Journal of Healthcare Engineering vol 1p 3972298
[4]. Alexander M Solomon B Ball D L et al. 2020 Evaluation of an artificial intelligence clinical trial matching system in Australian lung cancer patients JAMIA open vol 3 no 2 pp 209-215
[5]. Jianghua W et al. 2022 Artificial intelligence-assisted system for precision diagnosis of PD-L1 expression in non-small cell lung cancer Modern Pathology vol 35 no 3 pp 403-411
[6]. Li X Hu B Li H et al. 2019 Application of artificial intelligence in the diagnosis of multiple primary lung cancer Thoracic cancer vol 10 no 11 pp 2168-2174
[7]. Choi W Oh J H Riyahi S et al. 2018 Radiomics analysis of pulmonary nodules in low‐dose CT for early detection of lung cancer Medical physics vol 45 no 4 pp 1537-1549
[8]. Baldwin David R et al. 2020 External validation of a convolutional neural network artificial intelligence tool to predict malignancy in pulmonary nodules Thorax vol 75 no 4 pp 306-312
[9]. Huang P et al. 2019 Prediction of lung cancer risk at follow-up screening with low-dose CT: a training and validation study of a deep learning method The Lancet Digital Health vol 1 no 7 pp e353-e362
[10]. Akash N 2021 Lung Cancer Dataset Retrieved on 2024, Retrieved from: https://www.kaggle.com/datasets/akashnath29/lung-cancer-dataset/
Cite this article
Ji,A. (2024). Enhancing Lung Cancer Screening with Bidirectional LSTM and GRU Models. Applied and Computational Engineering,104,139-142.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 2nd International Conference on Machine Learning and Automation
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Chiu H Y Chao H S Chen Y M 2022 Application of artificial intelligence in lung cancer Cancers vol 14 no 6 p 1370
[2]. Kanan M Alharbi H Alotaibi N et al. 2024 AI-Driven Models for Diagnosing and Predicting Outcomes in Lung Cancer: A Systematic Review and Meta-Analysis Cancers vol 16 no 3 pp 674
[3]. Chen S 2022 Models of Artificial Intelligence‐Assisted Diagnosis of Lung Cancer Pathology Based on Deep Learning Algorithms Journal of Healthcare Engineering vol 1p 3972298
[4]. Alexander M Solomon B Ball D L et al. 2020 Evaluation of an artificial intelligence clinical trial matching system in Australian lung cancer patients JAMIA open vol 3 no 2 pp 209-215
[5]. Jianghua W et al. 2022 Artificial intelligence-assisted system for precision diagnosis of PD-L1 expression in non-small cell lung cancer Modern Pathology vol 35 no 3 pp 403-411
[6]. Li X Hu B Li H et al. 2019 Application of artificial intelligence in the diagnosis of multiple primary lung cancer Thoracic cancer vol 10 no 11 pp 2168-2174
[7]. Choi W Oh J H Riyahi S et al. 2018 Radiomics analysis of pulmonary nodules in low‐dose CT for early detection of lung cancer Medical physics vol 45 no 4 pp 1537-1549
[8]. Baldwin David R et al. 2020 External validation of a convolutional neural network artificial intelligence tool to predict malignancy in pulmonary nodules Thorax vol 75 no 4 pp 306-312
[9]. Huang P et al. 2019 Prediction of lung cancer risk at follow-up screening with low-dose CT: a training and validation study of a deep learning method The Lancet Digital Health vol 1 no 7 pp e353-e362
[10]. Akash N 2021 Lung Cancer Dataset Retrieved on 2024, Retrieved from: https://www.kaggle.com/datasets/akashnath29/lung-cancer-dataset/









