1. Introduction
Head tumors, including gliomas, meningiomas, and metastatic tumors, have high incidence and mortality rates globally, with rising trends each year. Early, accurate diagnosis is crucial for improving survival rates and quality of life. Traditional diagnostic methods in medical imaging, however, often rely on radiologists’ subjective judgment, leading to potential diagnostic uncertainty. The rapid advancement of deep learning, particularly convolutional neural networks (CNNs), has introduced transformative opportunities in medical imaging, significantly outperforming traditional methods in tasks such as image classification, segmentation, and detection [1].
ResUNet, a deep learning model combining residual learning and U-Net, was developed to enhance segmentation performance by preserving multi-level feature information through skip connections [2]. This architecture improves recognition of complex structures and strengthens robustness in medical imaging tasks. In head tumor diagnosis, ResUNet excels at capturing tumor shapes, boundaries, and relationships with surrounding tissues, offering more precise segmentation. By automating image analysis, ResUNet increases diagnostic efficiency and reduces human errors, such as misdiagnoses or missed diagnoses.
In the diagnosis of head tumors, ResUNet can better capture the shape, boundaries, and relationships of tumors with surrounding tissues, providing more accurate segmentation results. Through in-depth analysis of medical images, ResUNet can identify subtle lesions, helping doctors make more precise judgments in the early stages. Additionally, leveraging the automation capabilities of deep learning, ResUNet can not only improve diagnostic efficiency but also reduce misdiagnoses and missed diagnoses due to human factors. This paper aims to explore the effectiveness of ResUNet in head tumor diagnosis, analyzing its advantages and limitations, and providing theoretical and practical guidance for future clinical applications. Through in-depth research, we hope to provide new ideas and methods for the early diagnosis of head tumors, ultimately improving patient prognosis and quality of life.
2. Literature Survey
In recent years, Convolutional Neural Networks (CNNs), especially U-Net, have become central to medical image segmentation due to their strong performance. However, U-Net has limitations with non-standard shapes and variable imaging conditions, and thus integrating it with ResNet, as in ResUNet, has been a key advancement to enhance segmentation stability and generalization.
Zhang Qianwen et al [3]. applied 3D ResUNet to lung nodule segmentation by preprocessing CT scans, expanding them into 3D inputs, and generating data-augmented samples. Their model showed high recall, accurately filtering non-nodule regions, laying a solid foundation for true/false positive classification.
Yin Guangzhi et al [1]. developed the D2H-ResUNet model to generate high-quality hillshade maps from DEM data. By constructing a training set from hillshade-DEM pairs and applying ResUNet, their model produced visually superior maps with better adaptability across resolutions, enhancing the aesthetic and geomorphic representation of landscapes at different scales.
Similarly, Koteswara Rao Kodepogu et al [4]. applied ResUNet for liver and liver tumor segmentation on CT images. After finding U-Net’s performance lacking, ResUNet provided substantial improvement in segmentation efficiency and accuracy, proving more effective for complex tasks in medical imaging. Bai Penggang et al [5]. found that ResUNet also has certain advantages in a comparative studyof automatic organ segmentation in cervical cancer radiotherapy based on U-Net and ResUNet++models.
In 2024, Shuwan Feng, Ruixiang Song, and their team introduced an enhanced U-Net algorithm, incorporating SimAM and SE attention modules, to improve performance in remote sensing image segmentation [6]. This model, validated on remote sensing datasets, achieved a 17.41% and 13.23% improvement in MIoU, with MPA and accuracy gains of 16.88% and 13.045%, as well as 13.98% and 10.67%, respectively. The integration of SimAM and SE attention mechanisms led to remarkable advancements in both accuracy and visual clarity, showcasing robust generalization and resilience.
3. Brain-mri-detection-segmentation Based on Resunet
3.1. Datasets
This study utilizes the MRI brain tumor segmentation dataset provided by Kaggle to explore the application of deep learning techniques in medical image analysis. This dataset contains MRI images of multiple patients and precise annotation of their tumor regions, providing valuable resources for developing efficient segmentation algorithms. Validation_stplit=0.1, 10% is used to validate the dataset, and the remaining 90% is used for the training set. This is very important for applications that require high-precision images, such as medical imaging, map production, etc. This dataset is a medical imaging dataset used for brain tumor image segmentation, specifically for low-grade gliomas (LGG). These images are from The Cancer Imaging Archive (TCIA). They correspond to low-grade glioma (LGG) patients in The Cancer Genome Atlas (TCGA), which contains at least liquid attenuated inversion recovery (FLAIR) sequences and genomic cluster data.
3.2. Building a Classification Model and Train
In deep learning, theoretically the deeper the network structure, the more expressive the model should be, and the more complex the feature representation can be learned. However, in practice, as the network layers increase, the accuracy tends to saturate or even begin to decline, which is difficult to avoid even when non-saturating activation functions such as ReLU are used. This phenomenon is known as the network degradation problem. The previous view was that this was caused by overfitting, but later studies have found that it is difficult to solve this problem even by increasing the training data, and deeper networks do not show as much difficulty in training as they should, suggesting that it is not overfitting, but rather an optimization problem caused by a network that is too deep.
To solve the degeneracy problem, ResNet proposes a residual learning framework. The basic idea is that if the target mapping we want to learn is \( H(X) \) , we let the network fit a residual mapping. \( F(x)=H(x)-x \) Thus, the original target mapping becomes \( F(x)+x \) . This design allows the network to learn the residual mapping instead of directly learning the target mapping, which solves the problem of gradient vanishing and gradient exploding in the deep network, and improves the network's training efficiency and accuracy [7].
Therefore, I plan to apply transfer learning with the pretrained ResNet50 model for a brain tumor task, fine-tuning it on a new dataset. With frozen layers, I add custom layers (AveragePooling2D, Flatten, Dense, Dropout) to complete the model, leveraging existing knowledge to accelerate learning and enhance classification performance.
3.3. Building a Segmentation Model and Train
This study employs ResUNet, a hybrid deep learning model integrating ResNet and U-Net, for high-precision image segmentation. By embedding residual connections in U-Net’s encoder-decoder paths, ResUNet enhances feature learning, addresses gradient vanishing, and simplifies training. ResUNet maintains U-Net’s encoder-decoder framework to capture contextual information by progressively reducing spatial dimensions in the encoder and merging feature maps in the decoder.
The dataset was split into 85% training, 11.25% test, and 3.75% validation sets. The architecture comprises initial downsampling with convolutional layers, batch normalization, ReLU, and max pooling, followed by residual blocks in Stages 2–5 for deep feature extraction. The bottleneck stage refines features, and upsampling stages restore spatial resolution through upsampling, concatenation, and residual blocks. The final layer uses a 1x1 convolution with a sigmoid function to produce binary segmentation masks. Experimental results confirm ResUNet’s effectiveness in medical image segmentation tasks.
Table 2: Main hierarchy of the resunet
Layer(type) | Output shape | Parameters | Layer(type) | Output Shape | Parameters |
conv2d*2 | (None,256,256,16) | \ | batch_normalization | (None,256,256,16) | 64 |
MaxPool2D | (None,128,128,16) | 0 | ResBlock | (None,128,128,32) | \ |
MaxPool2D | (None,64,64,16) | 0 | ResBlock | (None,64,64,16) | \ |
MaxPool2D | (None,32,32,16) | 0 | ResBlock | (None,32,32,128) | \ |
ResBlock | (None,16,16,256) | \ | UpSampling2D | (None,32,32,256) | 0 |
Concatenate | (None,32,32,384) | 0 | ResBlock | (None,32,32,128) | \ |
UpSampling2D | (None,64,64,128) | 0 | Concatenate | (None,64,64,192) | 0 |
ResBlock | (None,64,64,64) | \ | UpSampling2D | (None,128,128,64) | 0 |
Concatenate | (None,128,128,96) | 0 | ResBlock | (None,128,128,32) | \ |
UpSampling2D | (None,256,256,32) | 0 | Concatenate | (None,256,256,48) | 0 |
conv2d*2 | (None,256,256,16) | \ | batch_normalization | (None,256,256,16) | 64 |
Add | (None,256,256,16) | 0 | Activation | (None,256,256,16) | 0 |
conv2d | (None,256,256,1) | 17 | Total | 1210513 |
I used the Adam optimizer during training, which combines adaptive gradient algorithm and root mean square propagation. It performs well in handling large-scale models by calculating the first-order and second-order moment estimates of gradients to adjust the learning rate of each parameter, thereby achieving more efficient network training. And the loss function focal_tversky, evaluation metric tversky, and callback function are set to ensure continuous learning improvement and automatic adjustment of learning rate.
4. Result Analysis
4.1. Loss Function and Accuracy
In this classification experiment, we use a loss function to measure the difference between the model's predicted values and the true values. Loss refers to quantifying the prediction error of a model into a non negative real number. The smaller the value of the loss function, the closer the prediction result of the model is to the true value. In this model, categorical_crossentropy is used as the loss function, and its formula is L= \( -\sum _{c=1}^{C}{y_{ic}}{log_{({\hat{y}_{ic}})}} \) . L is the loss value; \( {y_{ic}} \) is the value of the c-th category in the true label of sample i; \( {\hat{y}_{ic}} \) is the probability that the model predicts sample i belongs to the c-th category; log is a natural logarithm. Correspondingly, ACC is used to measure the accuracy of model predictions, which is defined as the proportion of successfully predicted samples to the total number of samples. The formula is ACC= \( \frac{number of correct predictions}{total number of predictions}×100\%. \)
First, for resnet50 (classification model evaluation of data sets), through historical research, we obtained that the loss of the LOSS and Acc value models reached 0.2353 and the Acc reached 94.75% after passing 50epochs. Then we called the pyplot package in matplotlib to record and visualize each training model.
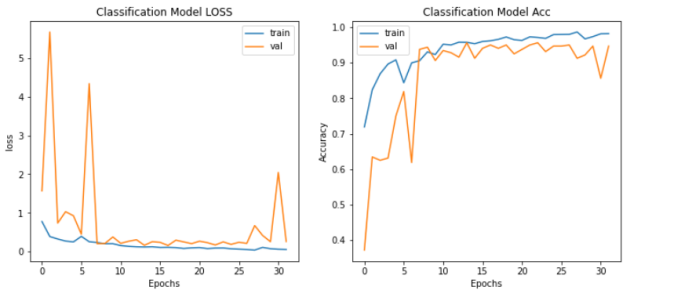
Figure 1: The scoring rate and loss value of the classification model
Secondly, for the segmentation model (reunet), and finally, I used the Matplotlib library to plot the losses and Tversky scores during the training and validation process. The Tversky function is a metric used to measure the similarity between two sets, defined as \( S(A,B)=\frac{|A∩B|}{∣A∩B∣+α∣A-B∣+β∣B-A∣} \) Among them, A and B are two sets |A-B| represents the number of elements in set A that are not in set B.| A-B | represents the number of elements in set A that are not in set B. ∣B − A∣represents the number of elements in set B that are not in A. α and β are nonnegative weight parameters used to control the influence of different set parts on similarity measurement. The segmentation accuracy of tversky's test set is 91.68%, with a loss value reduced to 0.1546. This indicates that the model can accurately segment tumor regions in images.
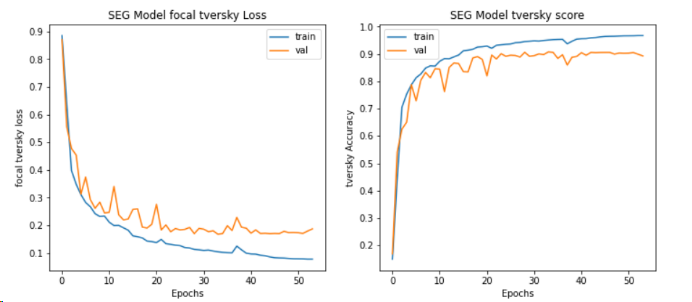
Figure 2: The tversky loss and tversky score of seg model
4.2. Visualizing Prediction
Visualize the prediction results of brain MRI images, including the original image, original mask, AI predicted mask, and the effect of overlaying the mask on the MRI image. From the figure, it can be seen that the segmented image can clearly display the location information of the brain tumor, which is basically the same as the position marked by the mask, and is marked with different colors and displayed together in the original image.
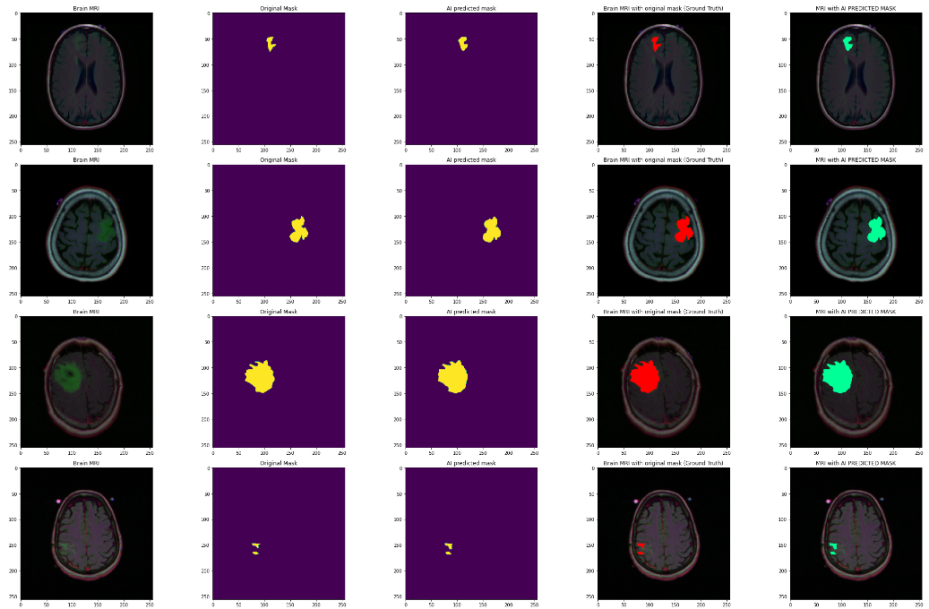
Figure 3: Partial images of brain tumors after segmentation
5. Conclusion
In this study, I used both ResNet50 and ResUNet to classify and segment head tumors simultaneously. For the Resnet50 model, residual learning is introduced to solve the gradient vanishing problem in deep network training.
In the classification problem, the loss value decreased to 0.2353 and the accuracy reached 0.9475, indicating that the model can effectively distinguish whether there is a tumor in the patient's head. In the test set, the Tversky loss value of Resunet is 0.9168, and the segmented Tversky reaches 91.68%. The results showed that ResNet50 combined with the Resunet model performed well in the MRI brain tumor segmentation dataset and could accurately identify brain tumors.
Overall, ResNet50 and ResUNet models are combined to segment brain tumors in magnetic resonance imaging (MRI), including the entire tumor, tumor core, and enhanced tumor regions. However, the data shows that the optimized model has high accuracy, and although there may be potential judgment errors, these results still demonstrate the potential of deep learning in medical image recognition. In subsequent research, using self supervised learning methods to extract richer features or adding pre trained base models for model fusion can help improve the accuracy of the model. In addition, considering the size of the dataset, more specialized labeled data can be collected in the future to test the model's generalization ability and further validate and enhance its robustness and applicability.
References
[1]. Yin Guangzhi, Li Shaomei, Ma Jingzhen, Lu Dongxu, & Bian Chenglin. A method for generating halo maps based on ResUNet. Journal of Wuhan University (Information Science Edition), 1-11. doi:10.13203/j.whugis20220532.
[2]. Su Fei, Wang Guanghui, Shi Yanxia, Jia Ran, Yan Yinfan, & Zu Linlu. (2024). Crop lesion segmentation model based on ResUnet with integrated channel attention mechanism. Journal of Chinese Agricultural Mechanization (08), 228-233. doi:10.13733/j.jcam.issn.2095-5553.2024.08.033.
[3]. Zhang Qianwen, Chen Ming, Qin Yufang, & Chen Xi. (2019). Lung nodule segmentation based on 3D ResUnet network. Chinese Journal of Medical Physics (11), 1356-1361.
[4]. Kodepogu, K. R., Muthineni, S. R., Kethineedi, C., Tejesh, J., & Uppalapati, J. S. (2023). Experimental Investigations to Detection of Liver Cancer Using ResUNet. Traitement du Signal, 40(5).
[5]. Bai Penggang, Wang Guohua, Chen Rongqin, Chen Jihong, Chen Wenjuan, Lin Jiafan & Ouyang Min. (2024). Comparison of Automatic Organ Segmentation Effects in Cervical Cancer Radiotherapy Based on UNet and ResUNet++ Models. Medical Equipment, (13), 1-6.
[6]. Feng, S., Song, R., Yang, S., et al. (2024). U-net Remote Sensing Image Segmentation Algorithm Based on Attention Mechanism Optimization. In 9th International Symposium on Computer and Information Processing Technology (ISCIPT), 633-636. IEEE.
[7]. Feng, S., Wang, J., Li, Z., et al. (2024). Research on Move-to-Escape Enhanced Dung Beetle Optimization and Its Applications. Biomimetics, 9(9), 517.
Cite this article
Deng,Y.;Cui,W.;Liu,X. (2024). Head Tumor Segmentation and Detection Based on Resunet. Applied and Computational Engineering,99,89-94.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 5th International Conference on Signal Processing and Machine Learning
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Yin Guangzhi, Li Shaomei, Ma Jingzhen, Lu Dongxu, & Bian Chenglin. A method for generating halo maps based on ResUNet. Journal of Wuhan University (Information Science Edition), 1-11. doi:10.13203/j.whugis20220532.
[2]. Su Fei, Wang Guanghui, Shi Yanxia, Jia Ran, Yan Yinfan, & Zu Linlu. (2024). Crop lesion segmentation model based on ResUnet with integrated channel attention mechanism. Journal of Chinese Agricultural Mechanization (08), 228-233. doi:10.13733/j.jcam.issn.2095-5553.2024.08.033.
[3]. Zhang Qianwen, Chen Ming, Qin Yufang, & Chen Xi. (2019). Lung nodule segmentation based on 3D ResUnet network. Chinese Journal of Medical Physics (11), 1356-1361.
[4]. Kodepogu, K. R., Muthineni, S. R., Kethineedi, C., Tejesh, J., & Uppalapati, J. S. (2023). Experimental Investigations to Detection of Liver Cancer Using ResUNet. Traitement du Signal, 40(5).
[5]. Bai Penggang, Wang Guohua, Chen Rongqin, Chen Jihong, Chen Wenjuan, Lin Jiafan & Ouyang Min. (2024). Comparison of Automatic Organ Segmentation Effects in Cervical Cancer Radiotherapy Based on UNet and ResUNet++ Models. Medical Equipment, (13), 1-6.
[6]. Feng, S., Song, R., Yang, S., et al. (2024). U-net Remote Sensing Image Segmentation Algorithm Based on Attention Mechanism Optimization. In 9th International Symposium on Computer and Information Processing Technology (ISCIPT), 633-636. IEEE.
[7]. Feng, S., Wang, J., Li, Z., et al. (2024). Research on Move-to-Escape Enhanced Dung Beetle Optimization and Its Applications. Biomimetics, 9(9), 517.









