1. Introduction
Cognition tasks are a key research domain in neuroscience which is related to memory, spatial representations, and generation. The function and investigation methods of two major glial cells, astrocytes and oligodendrocytes, which modulate memory have been discussed [1]. The more targeted investigations of human spatial schemas were also set up [2], while Jeff Hawkins proposed a new framework for understanding the neocortex [3]. However, the basic mechanism and the meaning of different biological functions in cognition are still ambiguous.
Meanwhile, Deep learning has been used in various areas and has resulted in many implementations like Generative Pre-trained Transformer (GPT), auto self-driving, and more. But compared to biology, there is still lots of potential to explore, like problems of energy consumption and generation. Hence, if deep learning has a fundamental connection to neuroscience, it will be promising to combine two domains to research astrocytes and oligodendrocytes. In the past ten years, many related works have implied potential for the connection. Andriy S. Kozlov added flexible mapping, which is inspired by biological neural networks to Convolutional Neural Networks (CNN), and found increased robustness to adversarial examples [4]. Computationally evidence involving Transformer was also attained, which investigates how humans extract meanings from language [5].
These studies show deep learning models can help interpret the mechanism of neuroscience. Meanwhile, from the level of neurons to the action mode of humans, neuroscience can also inspire the development of deep learning.
The contributions of this review focusing on the interaction between deep learning and biotic cognition elements are:
(1) From micro to macro, we summarize breakthroughs achieved by using deep learning in research of grid cells, neuron–astrocyte networks, and hippocampus.
(2) We classify neuroscience promotes deep learning in three directions: Modification of the raw models, Combination of different models, and raising of new models.
(3) Challenges that remained are discussed. How to select relevant biological features and translating them into algorithms and how to interpret the phenomenon of experiments to infer mechanisms are important.
(4) We conclude models used in these studies as biology-combined models (BCM). In the future, using BCM to implement spatial and non-spatial tasks will lead to more understanding of spatial encodes and generalizations. Combining neuroscience and deep learning may offer the potential to understand how neural networks work and increase both network’s interpretability. The cooperation of neuroscience researchers and AI scientists must be more diverse and scope-crossed.
2. Deep Learning Helps Interpret Neuroscience
2.1. Grid cells
Grid cells, initially found in the entorhinal cortex, play important roles in encoding space in navigation. Moreover, it is also considered to be essential in non-spatial tasks, like language and logic. However, the mechanisms of its spatial representations and generalization of related points in abstract tasks still leave some puzzles. In recent years, several works have offered new views of grid cells from deep learning.
2.1.1. RNN Interprets Spatial Representations
Based on velocity inputs, navigation tasks in 2D arenas are performed by recurrent neural networks (RNNs). It finds grid-like and various spatial correlated unit functions emerge in RNN. Not only types of neurons, but the sequence of emergence is also consistent with medical experiments. These results suggest that grid cells and other spatial units may represent an inherently efficient solution for space representation, particularly in the context of the predominant recurrent connections present within neural circuits [6].
2.1.2. DNN Interprets Grid Encoding
Grid coding provides an effective representational framework for both biological and artificial neural networks. Previous studies have demonstrated that memory-based deep neural networks, such as long short-term memory (LSTM), can replicate grid cell-like activity during speed-based path integration tasks in navigation. Li and colleagues further argue that grid-like firing patterns are more general, extending beyond navigation algorithms and network architectures. This assertion is supported by non-spatial experiments, which confirm that the formation of grid-like representations is driven by fundamental neural encoding mechanisms. Consequently, grid cells are considered a universal neural representation, applicable beyond spatial processing tasks [7].
2.1.3. Determinantal Point Process Attention Help Interpret Distribution Generalization
It is mysterious how the human brain supports complex forms of generalization. An algorithm is set up to achieve out-of-distribution (OOD) generalization. The algorithm based on grid cell code consists attentional mechanism — Determinantal Point Process (DPP), which ensures maximum sparseness in space coverage. The experimental results indicate that this model achieves OOD generalization performance successfully across various cognitive tasks. It implies that research combination of grid code and DDP similar mechanisms in the biological brain may help interpret generalization performance [8].
2.2. Neuron-astrocyte
Astrocytes, a type of glial cell, are thought to have tight relationships to cognitive processes, which involve the function of neurons. And neuron–astrocyte interactions are across many timescales and spatial scales to form feedback loops. However, the computational capabilities of neuron–astrocyte networks in the brain are still not clear. To investigate it, Leo Kozachkova et al. set up a model proposing that these biological systems can perform computations similar to Transformers [9].
The methodology involves constructing an artificial neuron–astrocyte network that mimics the core functions of a Transformer. Particularly focusing on tripartite synapses, which connect an astrocyte with a presynaptic and a postsynaptic neuron, see Fig.1. This trilateral structure using Random Feature Mapping to facilitates the normalization process essential for the Transformer’s self-attention mechanism. Leo employed theoretical derivations and simulations to demonstrate the correspondence between their biological model and AI architecture. They address the challenges of temporal and spatial nonlocality inherent in traditional Transformers by delineating distinct writing and reading phases within the network.
The conclusions assert that neuron–astrocyte networks can effectively approximate the computations performed by Transformers, offering a normative framework for understanding the biological basis of such computations. The model’s flexibility allows it to approximate any Transformer, potentially elucidating the prevalence of astrocytes across various brain regions and species.
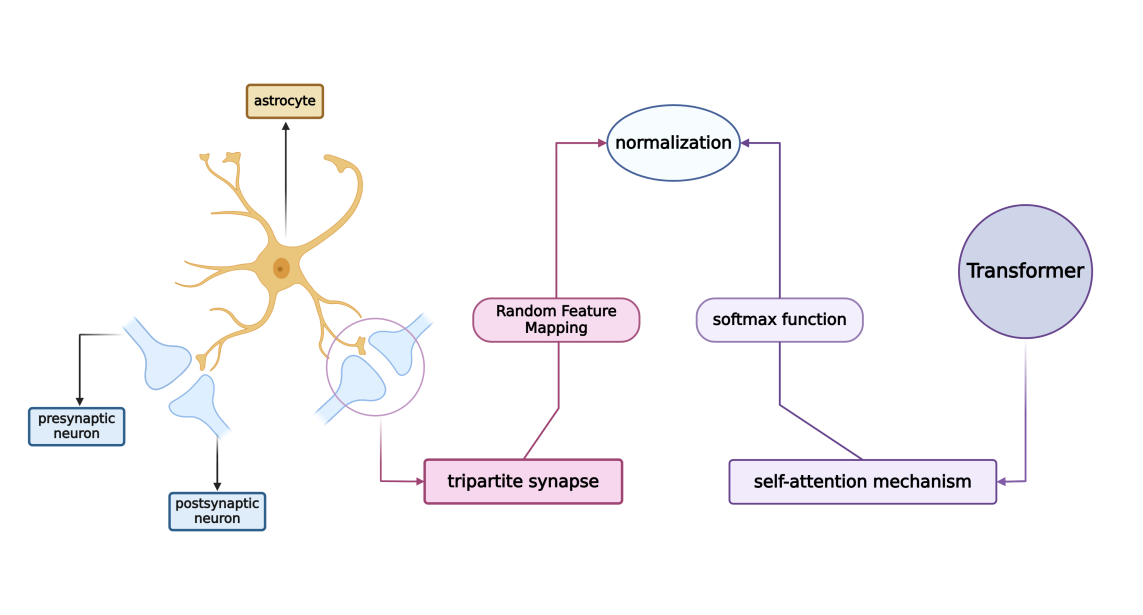
Figure 1. Transformer-based understanding of the astrocyte-neuron network.
2.3. Hippocampus
Spatial and relational memory tasks of the hippocampal-entorhinal system were the focus of James C.R. Whittington’s research. In 2020, a neuroscience model that captures many known neural phenomena in the hippocampus and entorhinal cortex was integrated into the Tolman-Eichenbaum Machine (TEM) by Whittington et al. [10]. Then, the relationship between transformers and TEM was analyzed by Whittington et al. [11].
The Temporal Encoding Model (TEM) elucidates the broader characteristics of the hippocampus by framing spatial and relational memory tasks as examples of structural abstraction and generalization. By linking limited structural representations to variable object perceptions, it demonstrates strong generalization capabilities applied to inferencing from changing perceptual information. One part of the model implements positioning and navigation, while the other part enables the mutual retrieval of object and location information.
The self-attention mechanisms in Transformers capture relationships by computing the interactions between Queries, Keys, and Values. In the TEM, memory retrieval is based on the dot product between queries and memory contents. Hence, the self-attention mechanisms in Transformers are similar to the memory retrieval components. Place cells in the hippocampus process spatial and sensory information for navigation. The sparse activation of the self-attention mechanism leads to neurons exhibiting spatial tuning similar to place cells. And path integration in Transformers can serve as positional encoding. This path integration is analogous to the function of place cells in biological systems, where the current position is calculated through path integration.
Transformers and TEM models may apply to brain regions beyond the hippocampus, particularly in cortical regions such as language areas. While transformers have been used to model the hippocampus and its connections, evidence shows that transformer representations also predict activity in language areas, and patients with significant hippocampal deficits can still engage in normal language function. Similar to spatial tasks, positional encodings in higher-order tasks such as language should reflect the abstract structure of the task (e.g. grammar). Therefore, dynamic positional encodings, inferred from learned structures, could be a productive research direction in domains such as language, mathematics, and logic.
3. Neuroscience Promotes Deep Learning Improvement
3.1. Modification of Models
By leveraging self-attention mechanisms, transformer-based models have impressive performance on NLP tasks and other sequential tasks [12]. However, the large order of magnitude and many dense matrices in the models lead to expensive training and inference. Thereby, some model pruning methods were proposed, while NeuroPrune provides a neuro-inspired approach to speed up both training and inference processes [13].
In the evolutionary process of the neuronal network, synapses are wisely removed after building over-abundant synapses, until the network gets stable. NeuroPrune mimics the synapse-cutting process, including preferential attachment and elimination of redundancy. As for preferential attachment, it is a concept also known as rich-get-richer that neurons with more connections build even more connections, while those with fewer connections are removed over time.
Inspired by preferential attachment, MLP layers, and attention heads will get sparsification. In the MLP layers, preferential attachment is achieved by adding a weighted \( {l_{1}} \) penalty to the training objective, where the weights for each row of entries in the matrix are inversely proportional to the (fractional) connectivity of that neuron. This means neurons with fewer connections are penalized more, encouraging a training process where sparsely connected neurons are likely to be weeded out. Essentially, preferential attachment makes the network sparser by focusing connections on already well-connected neurons, resulting in a more efficient and streamlined network.
Inspired by the elimination of redundancy, some similar heads will be removed. For attention heads, redundancy is eliminated by identifying and removing similar heads. The similarity between heads is measured using a distance threshold, and heads that are found to be similar are pruned to reduce redundancy. This process ensures that only the most unique and useful attention heads are kept, further optimizing the network’s performance and efficiency. This method draws parallels to how redundant synapses are eliminated in the brain, enhancing the network’s overall functionality by maintaining only the most critical connections.
3.2. Combination of Models
The TEM-transformer (TEM-t) model proposed by Whittington et al., improves the structure by incorporating mechanisms for processing spatial and sensory information found in biological systems. Specifically, using path integration as positional encoding enhances the Transformer’s ability to handle spatial information, allowing it to better simulate the brain’s functions in spatial navigation and memory.
The TEM-t model achieves spatial tuning like place cells through sparse activation and exhibits random remapping between different environments. This behavior is consistent with place cells in biological systems, indicating that introducing sparse activation and remapping mechanisms can improve the Transformer’s performance in handling multi-environment tasks.
3.3. A New Artificial Model: The Thousand Brains
The Thousand Brains Theory introduces a novel artificial neural network model inspired by the neocortex’s biological underpinnings. This model is based on the neocortex’s hierarchical structure, where grid cells and star-shaped astrocytes form a reference system essential for memory, learning, and cognition.
The neocortex, with its highly folded surface area, is the brain’s primary region for intelligence, and its operation is reflected in the HTM (Hierarchical Temporal Memory) model. The HTM leverages sparse distributed representations to encode information robustly and efficiently, akin to the neocortex’s energy-efficient processing [14].
This model supports online, continuous learning, unlike traditional AI, which is often batch-oriented and static. By incorporating the neocortex’s principles, including the dynamic interaction between grid cells for spatial mapping and the continuous prediction and learning from streaming sensory data, the HTM model advances the field of AI toward systems that can learn and adapt in a manner more closely aligned with human cognition.
This biologically constrained approach holds promise for developing AI with human-like cognitive capabilities, potentially revolutionizing the way machines understand and interact with the world. In the realm of artificial neural networks, the Thousand Brains Theory introduces a paradigm-shifting model that emulates the neocortex’s hierarchical structure and function. This model is distinguished by its reliance on sensory data, where each cortical column operates as an independent yet interconnected unit, processing information through a shared “language” — a universal algorithm that underpins cognitive capabilities [15].
The network’s architecture mirrors the neocortex’s columnar organization, with each column capable of autonomous problem-solving while contributing to collective intelligence. This design facilitates continuous online learning, akin to the neocortex’s ability to adapt in real time, and supports the formation of robust, sparse representations that are invariant to sensory input variations [16].
A pivotal feature of this model is the use of grid cells and displacement cells, which enable the network to construct comprehensive object models and understand the composition of complex structures. This common cortical algorithm not only enhances learning efficiency but also imbues the network with the flexibility to generalize knowledge across different domains, thus propelling the pursuit of artificial general intelligence.
4. Conclusion
Incorporating biological functions into algorithms or using algorithms to simulate some biological functions is a common theme in the works discussed above. For instance, by integrating grid codes and DDP to perform spatial and non-spatial tasks, researchers can use these models inspired by a certain biology function to test multiple tasks, even those previously considered irrelevant to that function. Therefore, using this kind of biology-combined model (BCM) can evaluate how much improvement artificial models achieve and find evidence of the mechanism of biology function.
However, future challenges have also been revealed in how to express biology models properly and infer mechanisms from experiments. To express biology models into algorithms is a key section in BCM and is full of uncertainty. Researchers must choose what kind of actions and features to add to deep learning, especially when the biological mechanisms are ambiguous and how deep learning works is also not clear. Most works mentioned in this review are developed on a solid theory foundation. When applying BCM in cutting-edge research, it needs more diversified evaluations to infer and verify results by collaboration of neuroscience researchers and AI scientists.
It is believed biological models that can inspire deep learning are gradually expanding, with room for exploration at each level: from individual neurons to neural networks, brain regions, the whole brain, and even organisms. The future deep learning may evolve towards more general, multimodal, and higher-level biologically-inspired models like a thousand brains even involving general sensor inputs. Biological models and algorithmic models have exhibited a relationship of mutual validation and understanding, which can promote collaborative optimization and result in a richer and more diverse framework. We predict future consensus will be more abundant.
Acknowledgments
Ziang Wei, Xiaoyu Liu, Tianxin Xie, Zoujizhong Wang, and Wanyue Bian contributed equally to this work and should be considered co-first authors.
References
[1]. A. Kol and I. Goshen, “The memory orchestra: the role of astrocytes and oligodendrocytes in parallel to neurons, ” Current Opinion in Neurobiology, vol. 67, pp. 131–137, Apr. 2021, doi: 10.1016/j.conb.2020.10.022.
[2]. D. Farzanfar, H. J. Spiers, M. Moscovitch, and R. S. Rosenbaum, “From cognitive maps to spatial schemas, ” Nat Rev Neurosci, vol. 24, no. 2, pp. 63–79, Feb. 2023, doi: 10.1038/s41583-022-00655-9.
[3]. J. Hawkins, M. Lewis, M. Klukas, S. Purdy, and S. Ahmad, “A Framework for Intelligence and Cortical Function Based on Grid Cells in the Neocortex, ” Front. Neural Circuits, vol. 12, p. 121, Jan. 2019, doi: 10.3389/fncir.2018.00121.
[4]. L. Evanson, M. Lavrov, I. Kharitonov, S. Lu, and A. S. Kozlov, “Biomimetic computations improve neural network robustness, ” Oct. 31, 2023. doi: 10.1101/2023.10.26.564127.
[5]. M. Schrimpf et al., “The neural architecture of language: Integrative modeling converges on predictive processing, ” Proc. Natl. Acad. Sci. U.S.A., vol. 118, no. 45, p. e2105646118, Nov. 2021, doi: 10.1073/pnas.2105646118.
[6]. C. J. Cueva and X.-X. Wei, “Emergence of grid-like representations by training recurrent neural networks to perform spatial localization, ” 2018, doi: 10.48550/ARXIV.1803.07770.
[7]. L. Songlin, D. Yangdong, and W. Zhihua, “Grid Cells Are Ubiquitous in Neural Networks, ” 2020, arXiv. doi: 10.48550/ARXIV.2003.03482.
[8]. S. S. Mondal, S. Frankland, T. W. Webb, and J. D. Cohen, “Determinantal point process attention over grid cell code supports out of distribution generalization, ” eLife, vol. 12, p. RP89911, Aug. 2024, doi: 10.7554/eLife.89911.3.
[9]. Kozachkov, L., Kastanenka, K. V., & Krotov, D. (2023). Building transformers from neurons and astrocytes. Proceedings of the National Academy of Sciences, 120(34).Available:https://doi.org/10.1073/pnas.2219150120
[10]. J. C. R. Whittington et al., “The Tolman-Eichenbaum Machine: Unifying Space and Relational Memory through Generalization in the Hippocampal Formation, ” Cell, vol. 183, no. 5, pp. 1249-1263.e23, Nov. 2020, doi: 10.1016/j.cell.2020.10.024.
[11]. J. C. R. Whittington, J. Warren, and T. E. J. Behrens, “Relating transformers to models and neural representations of the hippocampal formation, ” 2021, arXiv.doi:10.48550/ARXIV.2112.04035.
[12]. A. Vaswani et al., “Attention Is All You Need, ” Aug. 01, 2023, arXiv: arXiv:1706.03762. Accessed: Jul. 23, 2024. [Online]. Available: http://arxiv.org/abs/1706.03762
[13]. A. Dhurandhar et al., “NeuroPrune: A Neuro-inspired Topological Sparse Training Algorithm for Large Language Models, ” 2024, arXiv. doi: 10.48550/ARXIV.2404.01306.
[14]. K. J. Hole and S. Ahmad, “A thousand brains: toward biologically constrained AI, ” SN Appl. Sci., vol. 3, no. 8, p. 743, Aug. 2021, doi: 10.1007/s42452-021-04715-0.
[15]. M. Lewis, S. Purdy, S. Ahmad, and J. Hawkins, “Locations in the Neocortex: A Theory of Sensorimotor Object Recognition Using Cortical Grid Cells, ” Front. Neural Circuits, vol. 13, p. 22, Apr. 2019, doi: 10.3389/fncir.2019.00022.
[16]. J. Hawkins, S. Ahmad, and Y. Cui, “A Theory of How Columns in the Neocortex Enable Learning the Structure of the World, ” Front. Neural Circuits, vol. 11, p. 81, Oct. 2017, doi: 10.3389/fncir.2017.00081.
Cite this article
Wei,Z.;Liu,X.;Xie,T.;Wang,Z.;Bian,W. (2025). Explore Potential in Bridging of Neuroscience and Deep Learning. Applied and Computational Engineering,132,20-26.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 2nd International Conference on Machine Learning and Automation
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. A. Kol and I. Goshen, “The memory orchestra: the role of astrocytes and oligodendrocytes in parallel to neurons, ” Current Opinion in Neurobiology, vol. 67, pp. 131–137, Apr. 2021, doi: 10.1016/j.conb.2020.10.022.
[2]. D. Farzanfar, H. J. Spiers, M. Moscovitch, and R. S. Rosenbaum, “From cognitive maps to spatial schemas, ” Nat Rev Neurosci, vol. 24, no. 2, pp. 63–79, Feb. 2023, doi: 10.1038/s41583-022-00655-9.
[3]. J. Hawkins, M. Lewis, M. Klukas, S. Purdy, and S. Ahmad, “A Framework for Intelligence and Cortical Function Based on Grid Cells in the Neocortex, ” Front. Neural Circuits, vol. 12, p. 121, Jan. 2019, doi: 10.3389/fncir.2018.00121.
[4]. L. Evanson, M. Lavrov, I. Kharitonov, S. Lu, and A. S. Kozlov, “Biomimetic computations improve neural network robustness, ” Oct. 31, 2023. doi: 10.1101/2023.10.26.564127.
[5]. M. Schrimpf et al., “The neural architecture of language: Integrative modeling converges on predictive processing, ” Proc. Natl. Acad. Sci. U.S.A., vol. 118, no. 45, p. e2105646118, Nov. 2021, doi: 10.1073/pnas.2105646118.
[6]. C. J. Cueva and X.-X. Wei, “Emergence of grid-like representations by training recurrent neural networks to perform spatial localization, ” 2018, doi: 10.48550/ARXIV.1803.07770.
[7]. L. Songlin, D. Yangdong, and W. Zhihua, “Grid Cells Are Ubiquitous in Neural Networks, ” 2020, arXiv. doi: 10.48550/ARXIV.2003.03482.
[8]. S. S. Mondal, S. Frankland, T. W. Webb, and J. D. Cohen, “Determinantal point process attention over grid cell code supports out of distribution generalization, ” eLife, vol. 12, p. RP89911, Aug. 2024, doi: 10.7554/eLife.89911.3.
[9]. Kozachkov, L., Kastanenka, K. V., & Krotov, D. (2023). Building transformers from neurons and astrocytes. Proceedings of the National Academy of Sciences, 120(34).Available:https://doi.org/10.1073/pnas.2219150120
[10]. J. C. R. Whittington et al., “The Tolman-Eichenbaum Machine: Unifying Space and Relational Memory through Generalization in the Hippocampal Formation, ” Cell, vol. 183, no. 5, pp. 1249-1263.e23, Nov. 2020, doi: 10.1016/j.cell.2020.10.024.
[11]. J. C. R. Whittington, J. Warren, and T. E. J. Behrens, “Relating transformers to models and neural representations of the hippocampal formation, ” 2021, arXiv.doi:10.48550/ARXIV.2112.04035.
[12]. A. Vaswani et al., “Attention Is All You Need, ” Aug. 01, 2023, arXiv: arXiv:1706.03762. Accessed: Jul. 23, 2024. [Online]. Available: http://arxiv.org/abs/1706.03762
[13]. A. Dhurandhar et al., “NeuroPrune: A Neuro-inspired Topological Sparse Training Algorithm for Large Language Models, ” 2024, arXiv. doi: 10.48550/ARXIV.2404.01306.
[14]. K. J. Hole and S. Ahmad, “A thousand brains: toward biologically constrained AI, ” SN Appl. Sci., vol. 3, no. 8, p. 743, Aug. 2021, doi: 10.1007/s42452-021-04715-0.
[15]. M. Lewis, S. Purdy, S. Ahmad, and J. Hawkins, “Locations in the Neocortex: A Theory of Sensorimotor Object Recognition Using Cortical Grid Cells, ” Front. Neural Circuits, vol. 13, p. 22, Apr. 2019, doi: 10.3389/fncir.2019.00022.
[16]. J. Hawkins, S. Ahmad, and Y. Cui, “A Theory of How Columns in the Neocortex Enable Learning the Structure of the World, ” Front. Neural Circuits, vol. 11, p. 81, Oct. 2017, doi: 10.3389/fncir.2017.00081.









