1. Introduction
In recent years, growing public awareness of personal health has led to an increasing demand for convenient and trustworthy access to medical information. Many individuals, particularly those affected by visibly apparent conditions such as dermatological diseases, tend to seek help online due to the complexity, time constraints, and financial burden associated with traditional medical consultations. However, information retrieved through general search engines is often fragmented, poorly structured, and lacks credibility, making it difficult for users to obtain reliable health guidance.
Therefore, the core motivation of this work is to enhance the reliability of LLMs in the medical domain by grounding their responses in structured, verifiable knowledge. To achieve this, our research aims to answer the following key questions:
(RQ1) How can a high-quality, domain-specific knowledge graph be efficiently constructed to cover the essential aspects of dermatology?
(RQ2) How can relevant knowledge be accurately retrieved from the graph, even when user queries are ambiguous or use non-technical terms?
(RQ3) To what extent does augmenting an LLM with this knowledge graph-based retrieval system improve the factual accuracy and reduce the "hallucination" in its responses to dermatological questions?
To address these research questions, this study propose a retrieval-augmented question-answering framework enhanced with a structured dermatological knowledge graph. The methodological workflow of this study is shown in Figure 1.
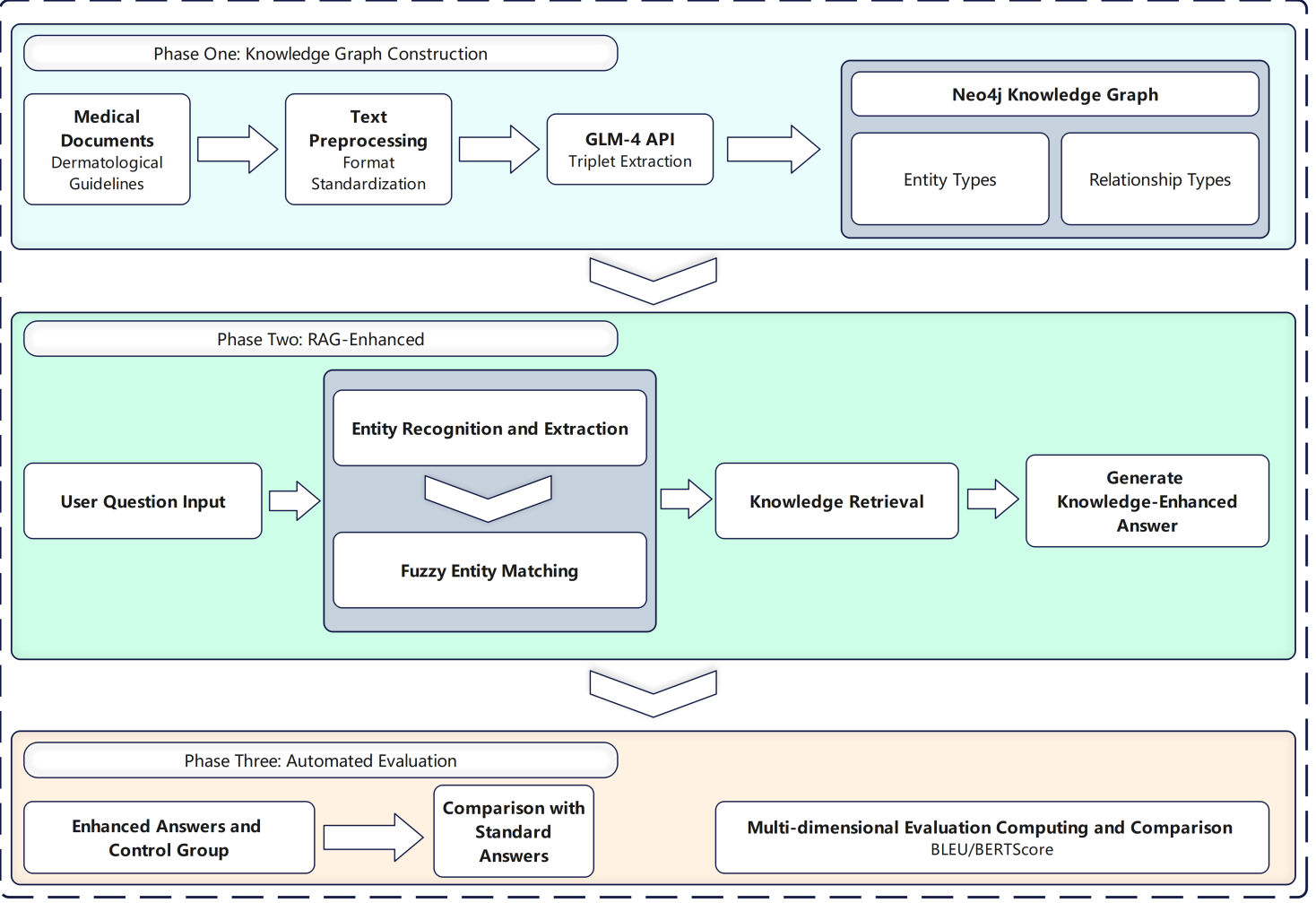
The key contributions, which directly correspond to these questions, include: (1) the construction of a dermatology-specific knowledge graph ontology with 19 key relation types (addresses RQ1); (2) an automated knowledge extraction method using GLM-4 to build the graph from authoritative medical sources (addresses RQ1); and (3) a fuzzy entity recognition mechanism to retrieve relevant knowledge and guide the LLM’s response generation (addresses RQ2). Experimental results demonstrate that our method significantly improves response accuracy and reduces hallucinations, providing a positive answer to RQ3 and offering a practical and scalable solution for safe and reliable medical AI applications.
The structure of this paper is organized as follows :Section 2 introduces related research work; Section 3 elaborates on our proposed knowledge graph construction and retrieval-augmented question-answering methods; Section 4 presents experimental results and analysis; and finally, Section summarizes the contributions of this paper and discusses directions for future research.
2. Related work
Advancements in Large Language Models:
Large Language Models (LLMs) represent a significant breakthrough in natural language processing, with their development traceable to the introduction of the Transformer architecture [1]. The early BERT model [2] pioneered deep bidirectional representations for pre-training, raising scores on the GLUE benchmark to 80.5%, substantially outperforming previous models. Subsequently, GPT-3 [3] expanded the parameter scale to 175 billion, demonstrating remarkable few-shot learning capabilities and achieving performance comparable to specially fine-tuned models across various NLP tasks.
In recent years, researchers have focused on enhancing the reliability and alignment of LLMs. Ouyang et al [4].trained models to follow human instructions using Reinforcement Learning from Human Feedback (RLHF), demonstrating that even the smaller-parameter InstructGPT model could outperform the larger GPT-3 in human preference evaluations. Nevertheless, LLMs still face "hallucination" issues in specialized domains such as healthcare—generating content that appears reasonable but is actually inaccurate. Kim et al [5].conducted a systematic analysis revealing that 91.8% of healthcare professionals had encountered these issues in practice, with 84.7% believing these hallucinations could impact patient health.
Current State of Medical Question-Answering Systems:
Research on medical question-answering systems has achieved significant progress in recent years. Liévin et al [6].found that the Codex model using Chain of Thought (CoT) prompting achieved an accuracy of 60.2% on the USMLE medical licensing examination, surpassing the passing threshold, while the Llama 2 70B model reached an even higher accuracy of 62.5%. Expert evaluations showed that InstructGPT demonstrated correct reasoning steps in 70% of cases, indicating that recent LLMs possess considerable medical reasoning capabilities.
The Med-PaLM 2 model developed by Singhal et al [7]. achieved 86.5% accuracy on USMLE-style questions through multiple optimization strategies, an improvement of over 19% compared to its predecessor. In real medical consultation environments, specialist physicians preferred the model's answers over those of general physicians in 65% of cases and considered the model's answers to have equivalent safety.
Specialized medical LLMs have also made important advances. Li et al.'s [8] ChatDoctor, which fine-tuned the LLaMA model on real patient-doctor dialogue datasets and incorporated external knowledge base retrieval, surpassed ChatGPT in BERTScore evaluation (precision: 0.8444 vs. 0.837). Zhang et al.'s [9] HuatuoGPT, which combined supervised fine-tuning with reinforcement learning based on AI feedback, performed exceptionally well across multiple Chinese medical benchmarks, achieving win rates of 52-58% against ChatGPT.
Nevertheless, existing models still face challenges in accuracy and explainability when handling specific domains such as dermatology. Dermatological diseases, characterized by diverse symptoms and complex manifestations, impose higher requirements on models' understanding of professional knowledge, prompting researchers to explore methods that combine structured knowledge with LLMs.
Knowledge Graph-Enhanced Language Models:
Knowledge graphs offer new approaches for improving the accuracy of LLMs. Ji et al.'s [10]comprehensive review summarized knowledge graph representation, acquisition, and application, establishing a theoretical foundation for the integration of knowledge graphs and LLMs. The Retrieval-Augmented Generation (RAG) framework proposed by Lewis et al [11]., which combines parametric and non-parametric memory, achieved significant results in open-domain question answering (Natural Questions: 44.5%, TriviaQA: 68.0%), substantially reducing hallucination phenomena.
In the field of medical knowledge graph construction, Ernst et al.'s [12] KnowLife system employed distant supervision and consistency reasoning methods to automatically extract biomedical knowledge from multiple textual sources, constructing a knowledge graph with an average precision of 93%. Li et al.'s [13] GLAME method enhanced the editing process of LLMs through knowledge graphs, improving performance on multi-hop reasoning problems by approximately 11% compared to the best baseline, demonstrating the effectiveness of knowledge graphs in constraining model generation.
Regarding the integration of medical knowledge and language models, Roy and Pan [14] explored various methods for injecting medical knowledge into BERT, finding that converting knowledge triplets into natural language as a second input to the model yielded the best results, achieving an F1 score of 91.81% in clinical relation extraction. Fei et al.'s [15] BioKGLM adopted a three-stage training paradigm to inject biomedical knowledge graph information into the BERT model, achieving an F1 score of 92.34% in biomedical entity recognition, significantly outperforming models without knowledge injection (90.41%).
Pan et al [16]. systematically reviewed the opportunities and challenges of combining LLMs with knowledge graphs, proposing an "explicit-parameterized knowledge hybrid expression" paradigm. They analyzed the bidirectional promotion pathways where LLMs facilitate KG construction and KGs enhance LLMs, providing a theoretical framework for the integrated development of knowledge engineering and AI.
3. Methodology
This study proposes a knowledge graph-based retrieval-augmented question answering framework specifically designed for the dermatology domain. The framework consists of three core modules: dermatological knowledge graph ontology design, automated knowledge graph construction, and retrieval-augmented question answering mechanism. This section elaborates on the methods and technical implementation of each module.
3.1. Dermatological knowledge graph ontology
This study first designed a specialized medical knowledge graph ontology for the dermatology domain to provide a unified formal representation of dermatological professional knowledge. This ontology structure systematically expresses various dimensions of dermatological knowledge and their intrinsic connections, ranging from disease definitions and clinical manifestations to diagnostic and treatment approaches.
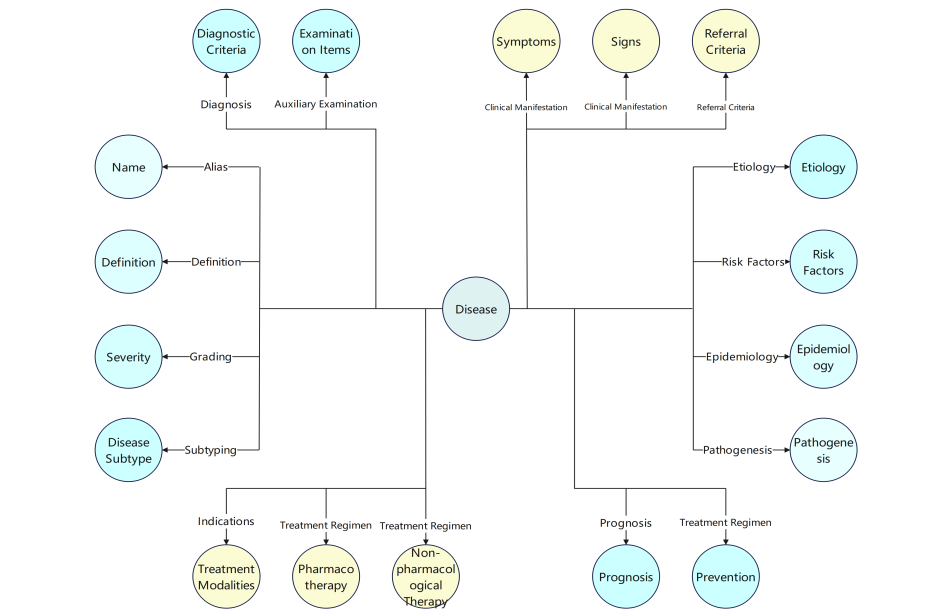
As shown in Figure 1, our dermatological knowledge graph ontology uses "Disease" as the central node, connecting to different types of medical entities through 19 key semantic relationships. These relationships include: definition, alias, etiology, risk factors, epidemiology, pathogenesis, clinical manifestations (symptoms and signs), classification, severity grading, auxiliary examinations, diagnosis, differential diagnosis, treatment plans (pharmacological and non-pharmacological treatments, preventive measures), prognosis, and referral indications. Additionally, we defined an "indication" relationship between "Treatment Method" and "Disease" to support clinical treatment decision-making.
3.2. Knowledge graph construction
3.2.1. Document processing
This study implemented a standardized medical document processing mechanism for correctly reading and parsing authoritative medical guideline documents. The system employs UTF-8 encoding to read documents, which is currently the most widely used text encoding standard capable of correctly handling specialized terminology and various special characters in medical literature. Through this unified document processing mechanism, this study can effectively extract content from different medical guidelines, providing a complete and accurate textual foundation for subsequent knowledge extraction. This step ensures the stability of the entire knowledge graph construction process and the integrity of the extracted knowledge.
3.2.2. Knowledge triplet extraction
We leveraged the powerful semantic understanding capabilities of the large language model GLM-4 to extract knowledge triplets conforming to the ontology structure from medical documents. In implementation, this study designed specific prompts for each relationship type, which clearly specified the head entity type, relationship type, and tail entity type to be extracted, and required the model to identify and extract corresponding knowledge instances from medical documents.
For example, for the "Disease-Symptoms-Clinical Manifestations" relationship, our designed prompt would explicitly guide the model to identify all symptomatic manifestations of a disease from the document. An example prompt is as follows:
“You are a medical knowledge graph construction expert.
Please extract only one specific type of knowledge triplet relationship about the specified disease from the following medical document.
Relationship type to extract:
- Head entity type: {head_type}
- Relation: {relation}
- Tail entity type: {tail_type}
For example, for the "Disease-Definition-Definition" relationship, you should extract the definition of psoriasis from the document.
Please extract all triplet instances that conform to the above relationship type from the following document:{document_text}”
Through this approach, the large language model can accurately identify and extract relevant entities and relationships from medical documents based on semantic understanding of specific relationship types, outputting formatted JSON results. These results contain complete triplet information, with each triplet comprising five components: head entity type, head entity, relationship, tail entity type, and tail entity.
3.2.3. Graph construction and storage
The extracted knowledge triplets were imported into a Neo4j graph database for storage and management. During the import process, this study first created corresponding node types for each ontology type, and then added extracted entities as nodes and relationships as connections between nodes to the graph database.
Specifically, this study used the py2neo library to connect to the Neo4j database and constructed the knowledge graph through the following steps: First, building the ontology structure by creating various types of nodes and their relationships; for each relationship type, calling the knowledge extraction module to obtain triplet data; creating head entities and tail entities as corresponding types of nodes; establishing specified types of relationships between head entity and tail entity nodes; using merge operations to ensure the uniqueness of nodes and relationships, avoiding duplication.
Through this approach, this study successfully constructed a dermatological knowledge graph containing 625 nodes and 19 types of relationships, providing structured knowledge support for subsequent question answering enhancement..
3.3. Retrieval-augmented question answering
The core innovation of this research lies in proposing a question answering enhancement paradigm based on knowledge graph retrieval, which significantly improves the accuracy of large language models in dermatological question answering through entity recognition, fuzzy matching, relationship extraction, and question augmentation.
3.3.1. Question key entity recognition
When a user poses a question, this study first utilize the large language model GLM-4-plus to identify key entities in the question. Unlike traditional named entity recognition methods, this study adopt a large language model-based entity type recognition approach that can simultaneously identify entities and their types. Specifically, this study use instructional prompts to guide the model to recognize specific types of entities from the question, such as diseases, symptoms, signs, etc., and output the results in the format of "entity type: entity name." The recognition results are then parsed into a list of (entity type, entity name) pairs, preparing for subsequent knowledge graph queries.
3.3.2. Entity fuzzy matchingecond section
Since user descriptions may differ from standard terminology in the knowledge graph, direct exact matching often leads to query failures. To address this issue, this study designed a large language model-based entity fuzzy matching mechanism.
The specific steps are as follows:
First, for each identified entity and its type, query all nodes of that type in the knowledge graph.
Second,end all possible candidate nodes along with the user-mentioned entity to the large language model.
Third,Through natural language prompts, have the model select the top 5 nodes most relevant to the user's description from the candidate nodes.
This method fully leverages the semantic understanding capabilities of large language models, capable of handling synonyms, near-synonyms, and expression variants, greatly improving the flexibility and accuracy of entity matching.
3.3.3. Knowledge retrieval and augmentation
For each successfully matched entity, this study extract related knowledge from the knowledge graph. Using Neo4j's Cypher query language, this study retrieve relationship paths within a maximum of two hops from the entity.
This graph-based query approach enables us to obtain knowledge that is not only directly related but also indirectly related, providing more comprehensive background information. For example, for the symptom "itching," we can not only obtain its corresponding diseases but also further retrieve related information such as treatment plans and diagnostic criteria for those diseases.
The retrieved paths are converted into knowledge statements in natural language description, in the form of "Entity A's relationship is Entity B." All these knowledge statements are aggregated as enhancement information for the original question..
4. Experiment results
4.1. Knowledge graph data statistics
This study successfully constructed a dermatological domain knowledge graph containing 625 nodes, covering 19 different types of entities. Table 2 details the quantity distribution of various types of nodes in the knowledge graph.
|
Node Type |
Count |
Node Type |
Count |
Node Type |
Count |
Node Type |
Count |
|
Disease |
12 |
Pathogenesis |
29 |
Disease Subtype |
24 |
Definition |
8 |
|
Symptom |
93 |
Epidemiology |
18 |
Pharmacological Treatment |
65 |
Alias |
13 |
|
Sign |
78 |
Diagnostic Criteria |
25 |
Non-pharmacological Treatment |
48 |
Referral Indications |
10 |
|
Risk Factor |
42 |
Examination Item |
31 |
Preventive Measures |
22 |
Treatment Method |
39 |
|
Etiology |
37 |
Severity |
15 |
Prognosis |
16 |
Total |
625 |
As shown in Table 2, symptom and sign nodes are the most numerous, with 93 and 78 respectively, reflecting the characteristic that dermatological diagnosis heavily relies on symptom and sign manifestations. This is followed by pharmacological treatment and non-pharmacological treatment nodes, with 65 and 48 respectively, demonstrating the diversity of treatment options for dermatological diseases. This distribution pattern aligns with the knowledge structure in the dermatology domain, providing comprehensive knowledge support for the question answering system.
4.2. Quantitative analysis
To objectively evaluate the performance of our proposed framework, this study conducted a quantitative analysis comparing the responses generated by the baseline LLM against those from our knowledge graph-enhanced framework. this study employed three standard automated evaluation metrics: BLEU and BERTScore. BLEU measures n-gram precision to assess fluency and adequacy, and BERTScore computes semantic similarity at the token level, which is crucial for evaluating factual correctness in domain-specific contexts. The aggregated results are presented in Table 3.
The results demonstrate a clear and consistent improvement achieved by our knowledge graph-enhanced framework.
BERTScore Improvement: The most significant gains are observed in the BERTScore metrics. The average Precision, Recall, and F1 scores increased from 0.5037, 0.7120, and 0.5897 to 0.5273, 0.7346, and 0.6135, respectively. The rise in Precision (P) indicates that the enhanced model's outputs are more factually correct and contain less irrelevant information. The increase in Recall (R) suggests that the responses successfully incorporate more key information from the reference answers. Consequently, the improved F1 score reflects a better overall balance of precision and recall, pointing to higher semantic quality.
BLEU Score Improvement: The average BLEU score also saw an increase from 0.0102 to 0.0157. While BLEU scores are typically low in open-ended question-answering tasks due to high lexical diversity, this relative improvement suggests that the enhanced responses share more precise keywords and phrases (n-grams) with the ground-truth answers.
In summary, the quantitative analysis provides strong evidence that augmenting the LLM with our dermatology knowledge graph leads to responses that are more accurate, comprehensive, and semantically aligned with expert-provided answers. These findings quantitatively validate the effectiveness of our framework in mitigating LLM "hallucinations" and improving response quality, thereby positively answering our third research question (RQ3).
|
Model |
BLEU |
BERTScore_P |
BERTScore_R |
BERTScore_F1 |
|
Ours |
0.0157 |
0.5273 |
0.7346 |
0.6135 |
|
Baseline |
0.0102 |
0.5037 |
0.7120 |
0.5897 |
5. Discussion
Our retrieval-augmented framework, which grounds LLM responses in a dermatology-specific knowledge graph, demonstrates a marked reduction in hallucination and a measurable improvement in answer accuracy compared to the baseline model. Quantitative metrics, such as increased BERTScore precision and recall as well as a higher BLEU score, indicate that the augmented responses include more relevant information and fewer factual errors. These findings confirm that integrating structured domain knowledge at inference time can effectively constrain LLM outputs in medical contexts and yield more reliable guidance.
The key advantage of our approach lies in its modular design: a finely defined ontology with nineteen relation types ensures comprehensive coverage of dermatological knowledge dimensions, and automated triplet extraction using a large language model aligns new data with the ontology schema without extensive manual curation. Additionally, the fuzzy entity recognition and matching mechanism, driven by LLM prompts, allows the system to interpret lay or variant expressions in user queries, thereby improving retrieval recall and robustness. By retrieving relevant graph-based facts at query time rather than embedding all knowledge into model parameters, the system remains adaptable to new guidelines or discoveries without necessitating costly model retraining.
Nevertheless, several limitations warrant careful consideration. The current knowledge graph, while covering core dermatological entities and relations, remains limited in scale and may omit rare conditions.Looking ahead, continuously expanding and validating the knowledge graph with new guidelines, research, and expert feedback, integrating multimodal data (e.g., clinical images), and enhancing transparency by citing retrieved evidence will strengthen reliability, while optimization of retrieval efficiency and personalized, context-aware responses will improve user experience; combined with rigorous evaluation using specialist-curated benchmarks and robust risk management, these efforts will ensure that grounding LLM-based QA in structured, domain-specific knowledge yields safe, accurate, and trustworthy medical guidance across specialties.
6. Conclusion
This study proposes a knowledge graph-based retrieval-augmented question answering framework to address the hallucination problem in large language models during dermatological consultations. By constructing a structured ontology with 19 semantic relationships, automating triplet extraction from medical guidelines using GLM-4, and integrating fuzzy entity recognition with knowledge-enhanced retrieval, our approach significantly improves the accuracy and reliability of responses in diagnosis, treatment, and prevention scenarios.
Despite its promising results, this work still faces limitations such as the absence of a large-scale evaluation dataset and the limited coverage of the knowledge graph. Future work will focus on building a dermatology-specific benchmark dataset, expanding the graph with diverse medical sources, and enhancing both knowledge extraction and entity matching algorithms. Ultimately, this study aim to generalize this framework across medical domains and modalities, contributing to more reliable, multimodal medical question answering systems that support clinical decisions and public health education.
References
[1]. Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A.N., Kaiser, Ł., Polosukhin, I. (2017) Attention Is All You Need. Advances in Neural Information Processing Systems, 30, 5998–6008.
[2]. Devlin, J., Chang, M. W., Lee, K., Toutanova, K. (2019) BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers), pp. 4171–4186.
[3]. Brown, T., Mann, B., Ryder, N., Subbiah, M., Kaplan, J., Dhariwal, P., Neelakantan, A., Shyam, P., Sastry, G., Askell, A., Agarwal, S., Herbert-Voss, A., Krueger, G., Henighan, T., Child, R., Ramesh, A., Ziegler, D.M., Wu, J., Winter, C., Hesse, C., Chen, M., Sigler, E., Litwin, M., Gray, S., Chess, B., Clark, J., Berner, C., McCandlish, S., Radford, A., Sutskever, I., Amodei, D. (2020) Language Models Are Few-Shot Learners. Advances in Neural Information Processing Systems, 33, 1877–1901.
[4]. Ouyang, L., Wu, J., Jiang, X., Almeida, D., Wainwright, C.L., Mishkin, P., Zhang, S., Agarwal, S., Slama, K., Ray, A., Schulman, J., Hilton, J., Kelton, F., Miller, L., Simens, M., Askell, A., Welinder, P., Christiano, P.F., Leike, J., Lowe, R. (2022) Training Language Models to Follow Instructions with Human Feedback. Advances in Neural Information Processing Systems, 35, 27730–27744.
[5]. Kim, Y., Jeong, H., Chen, S., Li, S.S., Lu, M., Alhamoud, K., Mun, J., Grau, C., Jung, M., Gameiro, R., Fan, L., Park, E., Lin, T., Yoon, J., Yoon, W., Sap, M., Tsvetkov, Y., Liang, P., Xu, X., Liu, X., McDuff, D., Lee, H., Park, H.W., Tulebaev, S., Breazeal, C. (2025) Medical Hallucinations in Foundation Models and Their Impact on Healthcare. arXiv preprint arXiv: 2503.05777.
[6]. Liévin, V., Hother, C.E., Motzfeldt, A.G., Winther, O. (2024) Can Large Language Models Reason about Medical Questions? Patterns, 5(3).
[7]. Rotmensch, M., Halpern, Y., Tlimat, A., Horng, S., Sontag, D.A. (2017) Learning a Health Knowledge Graph from Electronic Medical Records. Scientific Reports, 7(1), 5994.
[8]. Li, Y., Li, Z., Zhang, K., Dan, R., Jiang, S., Zhang, Y. (2023) ChatDoctor: A Medical Chat Model Fine-Tuned on a Large Language Model Meta-AI (LLaMA) Using Medical Domain Knowledge. Cureus, 15(6), e40895.
[9]. Zhang, H., Chen, J., Jiang, F., Yu, F., Chen, Z., Li, J., Chen, G., Wu, X., Zhang, Z., Xiao, Q., Wan, X., Wang, B., Li, H. (2023) HuatuoGPT, Towards Taming Language Model to Be a Doctor. arXiv preprint arXiv: 2305.15075.
[10]. Ji, S., Pan, S., Cambria, E., Marttinen, P., Yu, P.S. (2021) A Survey on Knowledge Graphs: Representation, Acquisition, and Applications. IEEE Transactions on Neural Networks and Learning Systems, 33(2), 494–514.
[11]. Lewis, P., Perez, E., Piktus, A., Petroni, F., Karpukhin, V., Goyal, N., Küttler, H., Lewis, M., Yih, W.-t., Rocktäschel, T., Riedel, S., Kiela, D. (2020) Retrieval-Augmented Generation for Knowledge-Intensive NLP Tasks. Advances in Neural Information Processing Systems, 33, 9459–9474.
[12]. Ernst, P., Siu, A., Weikum, G. (2015) KnowLife: A Versatile Approach for Constructing a Large Knowledge Graph for Biomedical Sciences. BMC Bioinformatics, 16, 1–13.
[13]. Li, D., Li, Z., Yang, Y., Zhang, X., Wang, H., Chen, M. (2024) Knowledge Graph-Enhanced Large Language Model for Domain-Specific Question Answering Systems. Authorea Preprints.
[14]. Roy, A., Pan, S. (2021) Incorporating Medical Knowledge in BERT for Clinical Relation Extraction. Proceedings of the 2021 Conference on Empirical Methods in Natural Language Processing, pp. 5357–5366.
[15]. Fei, H., Ren, Y., Zhang, Y., Liu, X., Wang, J., Zhao, Z. (2021) Enriching Contextualized Language Model from Knowledge Graph for Biomedical Information Extraction. Briefings in Bioinformatics, 22(3), bbaa110.
[16]. Pan, J.Z., Razniewski, S., Kalo, J.C., Nguyen, T., Smith, B. (2023) Large Language Models and Knowledge Graphs: Opportunities and Challenges. arXiv preprint arXiv: 2308.06374.
Cite this article
Wang,Z. (2025). Retrieval-Augmented Question Answering System Based on Large Language Models and Knowledge Graphs. Applied and Computational Engineering,178,236-245.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of CONF-CDS 2025 Symposium: Data Visualization Methods for Evaluatio
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A.N., Kaiser, Ł., Polosukhin, I. (2017) Attention Is All You Need. Advances in Neural Information Processing Systems, 30, 5998–6008.
[2]. Devlin, J., Chang, M. W., Lee, K., Toutanova, K. (2019) BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers), pp. 4171–4186.
[3]. Brown, T., Mann, B., Ryder, N., Subbiah, M., Kaplan, J., Dhariwal, P., Neelakantan, A., Shyam, P., Sastry, G., Askell, A., Agarwal, S., Herbert-Voss, A., Krueger, G., Henighan, T., Child, R., Ramesh, A., Ziegler, D.M., Wu, J., Winter, C., Hesse, C., Chen, M., Sigler, E., Litwin, M., Gray, S., Chess, B., Clark, J., Berner, C., McCandlish, S., Radford, A., Sutskever, I., Amodei, D. (2020) Language Models Are Few-Shot Learners. Advances in Neural Information Processing Systems, 33, 1877–1901.
[4]. Ouyang, L., Wu, J., Jiang, X., Almeida, D., Wainwright, C.L., Mishkin, P., Zhang, S., Agarwal, S., Slama, K., Ray, A., Schulman, J., Hilton, J., Kelton, F., Miller, L., Simens, M., Askell, A., Welinder, P., Christiano, P.F., Leike, J., Lowe, R. (2022) Training Language Models to Follow Instructions with Human Feedback. Advances in Neural Information Processing Systems, 35, 27730–27744.
[5]. Kim, Y., Jeong, H., Chen, S., Li, S.S., Lu, M., Alhamoud, K., Mun, J., Grau, C., Jung, M., Gameiro, R., Fan, L., Park, E., Lin, T., Yoon, J., Yoon, W., Sap, M., Tsvetkov, Y., Liang, P., Xu, X., Liu, X., McDuff, D., Lee, H., Park, H.W., Tulebaev, S., Breazeal, C. (2025) Medical Hallucinations in Foundation Models and Their Impact on Healthcare. arXiv preprint arXiv: 2503.05777.
[6]. Liévin, V., Hother, C.E., Motzfeldt, A.G., Winther, O. (2024) Can Large Language Models Reason about Medical Questions? Patterns, 5(3).
[7]. Rotmensch, M., Halpern, Y., Tlimat, A., Horng, S., Sontag, D.A. (2017) Learning a Health Knowledge Graph from Electronic Medical Records. Scientific Reports, 7(1), 5994.
[8]. Li, Y., Li, Z., Zhang, K., Dan, R., Jiang, S., Zhang, Y. (2023) ChatDoctor: A Medical Chat Model Fine-Tuned on a Large Language Model Meta-AI (LLaMA) Using Medical Domain Knowledge. Cureus, 15(6), e40895.
[9]. Zhang, H., Chen, J., Jiang, F., Yu, F., Chen, Z., Li, J., Chen, G., Wu, X., Zhang, Z., Xiao, Q., Wan, X., Wang, B., Li, H. (2023) HuatuoGPT, Towards Taming Language Model to Be a Doctor. arXiv preprint arXiv: 2305.15075.
[10]. Ji, S., Pan, S., Cambria, E., Marttinen, P., Yu, P.S. (2021) A Survey on Knowledge Graphs: Representation, Acquisition, and Applications. IEEE Transactions on Neural Networks and Learning Systems, 33(2), 494–514.
[11]. Lewis, P., Perez, E., Piktus, A., Petroni, F., Karpukhin, V., Goyal, N., Küttler, H., Lewis, M., Yih, W.-t., Rocktäschel, T., Riedel, S., Kiela, D. (2020) Retrieval-Augmented Generation for Knowledge-Intensive NLP Tasks. Advances in Neural Information Processing Systems, 33, 9459–9474.
[12]. Ernst, P., Siu, A., Weikum, G. (2015) KnowLife: A Versatile Approach for Constructing a Large Knowledge Graph for Biomedical Sciences. BMC Bioinformatics, 16, 1–13.
[13]. Li, D., Li, Z., Yang, Y., Zhang, X., Wang, H., Chen, M. (2024) Knowledge Graph-Enhanced Large Language Model for Domain-Specific Question Answering Systems. Authorea Preprints.
[14]. Roy, A., Pan, S. (2021) Incorporating Medical Knowledge in BERT for Clinical Relation Extraction. Proceedings of the 2021 Conference on Empirical Methods in Natural Language Processing, pp. 5357–5366.
[15]. Fei, H., Ren, Y., Zhang, Y., Liu, X., Wang, J., Zhao, Z. (2021) Enriching Contextualized Language Model from Knowledge Graph for Biomedical Information Extraction. Briefings in Bioinformatics, 22(3), bbaa110.
[16]. Pan, J.Z., Razniewski, S., Kalo, J.C., Nguyen, T., Smith, B. (2023) Large Language Models and Knowledge Graphs: Opportunities and Challenges. arXiv preprint arXiv: 2308.06374.









