1. Introduction
Cancer has been one major threat to human public health as the second‐leading cause of mortality in the United States. In 2024, an estimated 2,001,140 new cases of cancer will be diagnosed, and 611,720 deaths will occur from the disease in the United States and the world [1-4]. Cancer is characterized by a series of genetic alterations and metabolic changes that results in abnormal cell growth and proliferation [5]. Prevalent methods for cancer treatment include surgical removement, chemotherapy [6-9], and radiation therapy [10-11]; there are also emerging modalities including immunotherapy [12-14], phototherapy [15-17], and targeted therapies [18-19]. However, the majority of patients who receive these treatment suffer severe side effects and even relapses[6,20]. For instance, small molecules for chemotherapy can lead to hair loss, nausea, vomiting, diarrhea in patients, as they not only affect cancer cells but also kill normal healthy cells that proliferate rapidly. The repeated dosing resulting from the fast renal clearance and poor accumulation in tumors of such drugs further burdens the patients [21,22]. Therefore, developing drugs that have the capacity to actively target cancer cells may relieve the side effects and improve the life quality of cancer patients.
As one of the most prevalent treatments, chemotherapy covers around 50% of the patients worldwide. Chemodrugs disrupt the cellular functions within cancer cells in different ways. Alkylating drugs (e.g., cisplatin, dacarbazine) add alkyl groups to DNA during difference phases of cell division and replication [23]. Antimetabolites disrupt the metabolic activities such as inhibiting essential enzymes and interfering RNA translation or transcription [9]. Topoisomerase inhibitors bind to enzymes essential for DNA replications [24,25]. These activities lead to DNA breaks, impaired enzymatic functions, and reduced RNA and protein productions, which eventually result in cell arrest and apoptosis. By the mechanism of chemodrugs, cells that undergo high rate of growth and metabolism are to be impacted significantly, and thus cancer cells are sensitive to such treatment due to their extremely high cell proliferation and metabolic activity. However, healthy cells, such as hair follicle stem cells and intestinal epithelial cells, are also frequently renewed and thus affected during chemotherapy. In this regard, endowing the chemodrugs with the ability to enhance targeting cancer cells emerges as a research endeavor with fundamental potentials for treating cancer [26-29].
Endogenous machineries have been employed for cancer-targeting drug delivery. Low density lipoprotein, known as LDL, is natural carrier for lipids in our cells [30-32]. It is one of the five major groups of lipoprotein that transport all fat molecules around the body. LDL has a highly hydrophobic core consisting of polyunsaturated fatty acid known as oleates and hundreds to thousands of esterified and unesterified cholesterol molecules (Figure 1). This core also carries varying numbers of triglycerides along with other fats and is surrounded by a shell of phospholipids with unesterified cholesterol and a single copy of Apo B-100 [33]. Interestingly, LDL is highly positively associated with cancer development. Epidemiological studies have demonstrated that low-density lipoprotein (LDL) is closely associated with breast cancer, colorectal cancer, melanoma, leukemia, and other malignancies, suggesting that LDL plays important roles during the occurrence and development of cancers [31]. Intuitively, cancer cells demand a significant amount of fat and lipid molecules for the fast proliferation and cell growth. Several pieces of evidence indicate that various cancers upregulate the LDL receptor (LDLR) on the cell surface to scavenge LDL droplets. This inspired us that we can exploit the LDL as an active targeting carrier for hydrophilic chemodrugs and enhance the therapeutic outcomes.
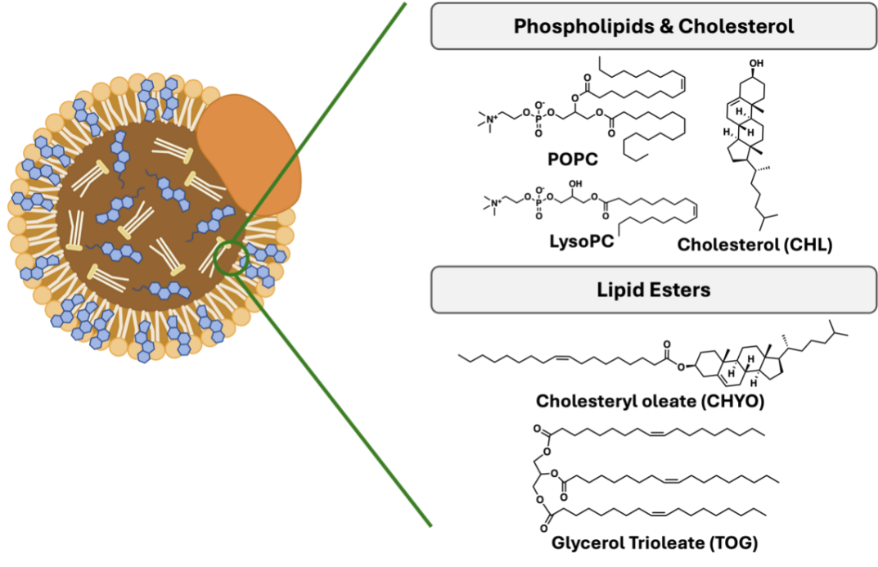
Figure 1: A schematic showing the shape and the chemical composition of an LDL droplet. It consists of an Apo-B protein on the surface of a lipid-based particle.
The question then becomes how we can engineer hydrophilic chemodrugs to allow them to partition into the LDL particles. Since the interior of LDL has a hydrophobic environment, we hypothesize that conjugating a hydrophobic moiety to the hydrophilic drugs with a reversible linkage may reverse the hydrophobicity of the drug and result in the favorable partition into LDL, which can subsequently carry the drugs to tumor sites and cells. Squalene emerges as a potential candidate due to its biocompatibility and readily availability. It is a natural 30-carbon triterpene and intermediate metabolite in the synthesis of cholesterol [34].
To confirm our hypothesis that squalene conjugation can enhance the partition of chemodrugs into LDLs, we used molecular modeling and coarse grained (CG) molecular dynamics (MD) simulations to simulate the partition process and quantify the associated free energy of free drugs and their squalene conjugates. As a proof of concept, we chose two model drugs, cladribine (CLA) and dacarbazine (DAC), and added two squalene moiety to form the conjugated drugs. We first successfully established the first CG LDL bilayer model that can be readily employed for a diverse simulation related to LDL droplets. From the simulations, we found that hydrophilic drugs possessed positive partition free energies while the squalene conjugates reversed the free energy profiles, exhibiting strong negative partition free energies, indicating that the partition of squalene conjugated drugs are strongly favored while that of the free drugs are disfavored. This evidence confirms our hypothesis that squalene conjugation can aid the chemodrugs to passively enter the LDL droplets due to the enhanced hydrophobic interactions with the LDLs. This partition, on one side, is likely to enhance the circulation of the chemodrugs, and on the other, will potentially enhance the drug accumulation because of the LDL targeting ability to cancers.
2. Simulation Methods
2.1. LDL Model Establishment
The size and complex components of LDL particles forbid us to model the whole particle. Therefore, we created and simulated a simplified model which contained ~10% of the full volume [35]. We used the following approximations to make the simulations tractable with the available computational resources while retaining the realistic lipid compositions and all major physicochemical ingredients of the LDL droplets. First, we excluded the surface Apo-B protein due to the fact that the interaction between the interior core of LDL particles with squalene-conjugated drugs presented the major event during the simulations. In addition, instead of simulating a whole spherical structure, we constructed a slice of the LDL particle and placed it in a periodic box as a bilayer model (Figure 2).The composition of each component is listed in Figure 2, in accordance with a realistic LDL particle and a previously reported coarse-grained model of LDL. The hydrophobic core includes cholesterol, glyceryl trioleates, and cholesteryl oleates as a random mixture. The two sides of the surface contain POPC, LysoPC aligned with their hydrophobic tails pointing to the core, and hydrophilic head to the water phase. A portion of cholesterols are added to each of the surface lipid regions to enhance the rigidity of the phospholipid layers.
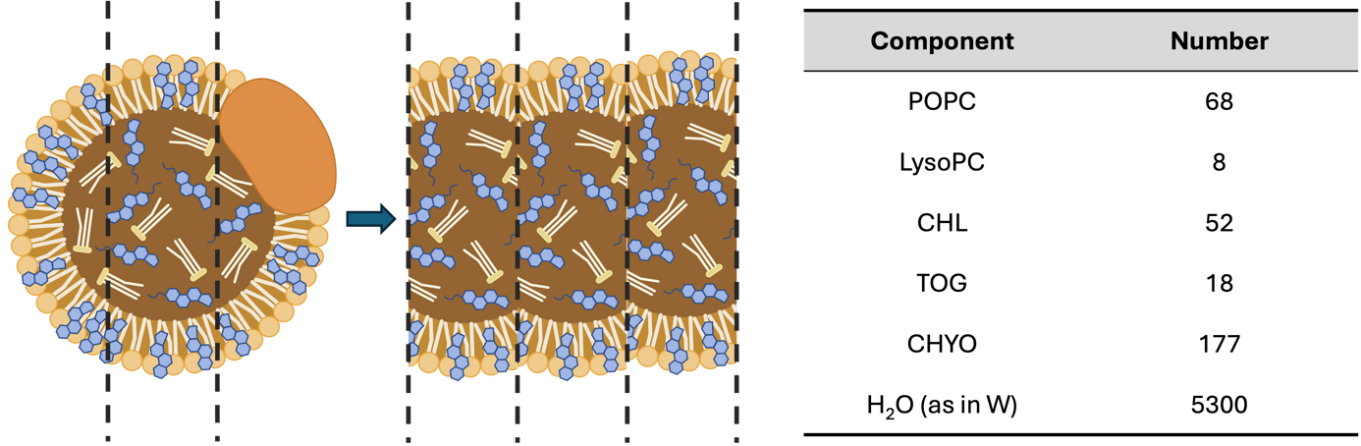
Figure 2: The schematic showing the establishment of the LDL model and the compositions in a single simulation box. The whole LDL droplet is sliced and modeled in a box with periodic boundary conditions as a bilayer model. The composition is around 10% of a whole volume of an LDL droplet.
We employed a coarse-grained (CG) modeling in order to reach the length scales necessary to model a substantial patch of the LDL and the time scales needed to converge the free energy calculations. The Martini model models a group of atoms (3-5) as a united bead, which reduces the simulation particles and permits us to implement simulations with a large time step [36]. Meanwhile, it possesses good description of biomolecular interactions and superior computation efficiency compared with all-atom simulations. All the topology files and force field parameters were obtained from the Martini database (https://mad.ibcp.fr) and assembled using packmol [37] into a box with a XYZ dimensions of 5×5×36 nm as the initial configuration. The initial configuration is shown in the Figure 2A.
2.2. Equilibration of the LDL Model
Prior to the modeling of the drugs and LDL, a pre-equilibration of the LDL model is required to converge the potential energy and other quantities for subsequent steered MD simulations and free energy calculations. MD simulations were carried out with the GROMACS 2023.1[38] package with periodic boundary conditions in three dimensions under the isothermal and isobaric ensemble. The temperature was maintained at 310 K by the langevin thermostat [39] while the pressure was regulated to be 1 bar using Berendsen barostat [40] for equilibration and c-rescale [41] barostat for sampling. The stochastic integrator was used to integrate the equations of motion for the Langevin dynamics with a default friction constant (γ) of 0.5 ps-1. The time step was set to 0.02 ps. Since the model is a bilayer model, we employed a semi-isotropic pressure coupling, which is a standard practice for simulating bilayer models. At each time step, the translational center-of-mass motion was removed. The Lennard-Jones potentials for van der Waals interactions were shifted to zero beyond a cutoff distance of 1.1 nm, while for Coulomb interactions, the reaction-field approach was used with a cutoff of 1.1 nm and εr = 15 (water). All the MD parameters were employed and maintained in accordance with the mdp files on the Martini official website.
For energy minimization, the steepest descent algorithms were used to eliminate any forces over 1000 kJ/mol/nm2 in the system. Then a 10 ns of NVT equilibration and a 10 ns NPT equilibration were conducted followed by 100 ns production run until we observe convergence on system potential energy and other related quantities. The final configuration was extracted and utilized as the starting configuration for studying the interaction between LDL and drug molecules. The potential energy profiles, density, and system temperature were extracted by gmx energy, while the partial density profiles were calculated using gmx density. All visualizations of the MD simulated configurations were realized by VMD [42].
2.3. Modeling the Drug Molecules
As a proof of concept, we selected two types of hydrophilic chemotherapy drugs, cladribine and dacarbazine, to confirm our hypothesis. Cladribine is an FDA approved chemotherapy drug for hairy cell leukemia which exhibits a characteristic of enhanced expression of LDLR [43]. Dacarbazine belongs to the alkylating chemodrugs used in clinics for the treatment of cancer of the lymph system and malignant melanoma, also with evidence of improved LDLR. Since the Martini database does model the small molecules, we parameterized the drug using an automated algorithm cg_param [44] which is reported to well describe the dynamics of small molecules. In Figure 3, we show the chemical structures and the AA to CG mapping for the two free drugs and their squalene conjugates. We appreciate that the cg_param program may be limited for complex drug models but we do anticipate that the distinct features of the drug and squalene can be reasonably represented to yield reasonable trends.
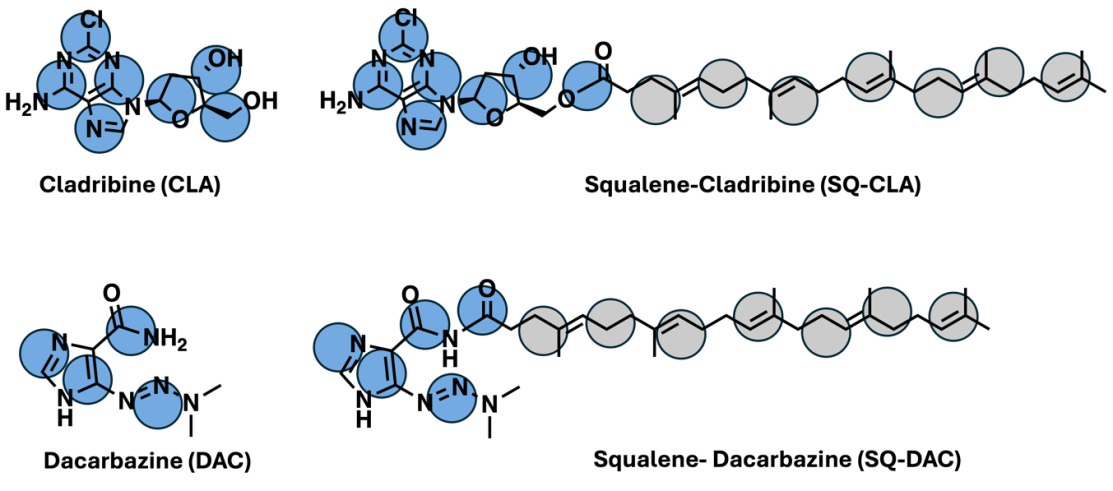
Figure 3: Beads representation of the free drug and squalene-conjugated drugs for cladribine and dacarbazine. The blue beads represent the free drugs, and the gray beads represent the squalene tails. Each bead was assigned to a bead type by cg_param algorithm [44]. The chemical structures were created by ChemDraw.
2.4. Steered MD Simulations
For computing the partition free energy using an alchemical transfer approach, we need a system where the drug resides in the LDL core as the starting configuration. To achieve this, we inserted one drug molecule into the upper water phase and generated 4 independent systems with each containing one type of drug. The initial position was determined to be ~ 3 nm above the LDL model, and a short NPT simulation of 1 ns was implemented for each system to equilibrate the water beads around the drug molecule. At the meantime, the drug was tethered to a harmonic spring with a force constant of 5000 kJ/mol/nm2 to prevent it drifting near the LDL surface. A long simulation may allow us to acquire configurations where the drug is inside the LDL core but can be intractable within our computation capacity. Thus, the steered MD simulations was utilized to pull the drug molecule into the LDL along the Z axis. The SMD simulations were performed under the NPT conditions over a course of 1.8 ns with a pulling rate of -0.005 nm/ps. The force constant of 3000 kJ/mol/nm2 was used. The COM distance between LDL and the drug was within 2 nm in the final configuration.
2.5. Free Energy Calculation by Alchemical Transfer Method
To compute the free energy associated to the partition of drug molecule into the LDL core (or reversibly the extraction of the drug molecule from LDL core to water phase), we employed an alchemical transfer protocol. Briefly, we copied the drug in the final configuration from the SMD simulation to the water phase. The drug in the LDL core is termed as MOL_1 while the one in water phase is denoted as MOL_2. For the initial state, MOL_1 is interacting with environment while MOL_2 is totally decoupled and noninteracting. Then, along the alchemical path, we simultaneously switch on the MOL_2 interactions and switch off the MOL_1 interaction, controlled by a parameter λ [45]. To prevent molecule drifting around, we applied a harmonic potential with a force constant of 1500 kJ/mol/nm2 to both molecules.
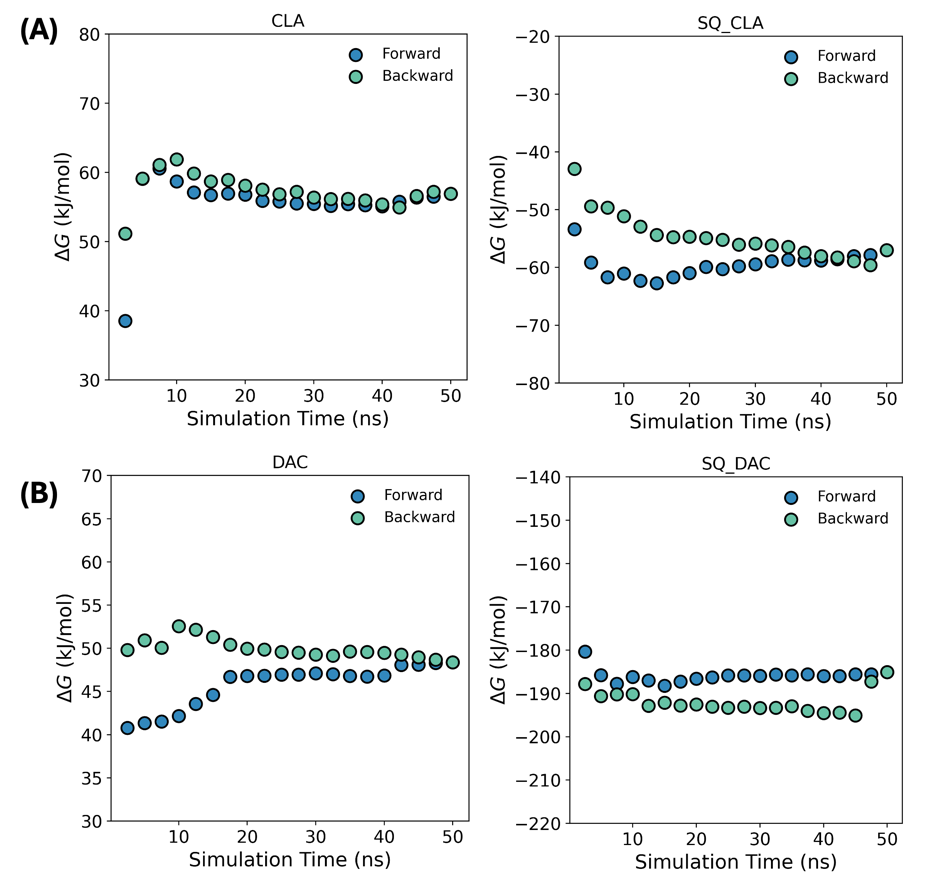
Figure 4: Convergence of the free energy calculation from different blocks of simulation time. All calculations have converged within the sampled time.
With an intermediate state λ, the Hamiltonian is described as
\( U(λ)=(1-λ){U_{0}}+λ{U_{1}} \)
in which \( {U_{0}} \) and \( {U_{1}} \) are the potential energy associated with the initial state and final state [46], and \( λ \) is the fictitious parameter from 0 to 1 linking the initial and final states. The free energy between the two states is then computed using the algorithm of gmx bar [47]. The time convergence of the free energy is calculated using alchemlyb [48,49] as shown in Figure 4.
3. Results
3.1. The Slice Model Retains the Properties of an LDL Particle
Prior to modeling the interaction between drugs and LDL particles, it is essential to obtain a well-equilibrated LDL model that is computationally tractable but also captures the property of a real LDL particle. Therefore, we established a slice model according to the previous report [35,50] using a Martini model by placing the esters within the core and phospholipid on the shell randomly (Figure 5A). After equilibration, we observe that the model shrinks due interaction between the molecules. To check equilibration process, we monitored thermodynamic quantities including potential energy, temperature, and density during the equilibration simulation. In Figure 5B-D, all these quantities are shown to have converged within the simulation time indicating that the system is stable and has well equilibrated.
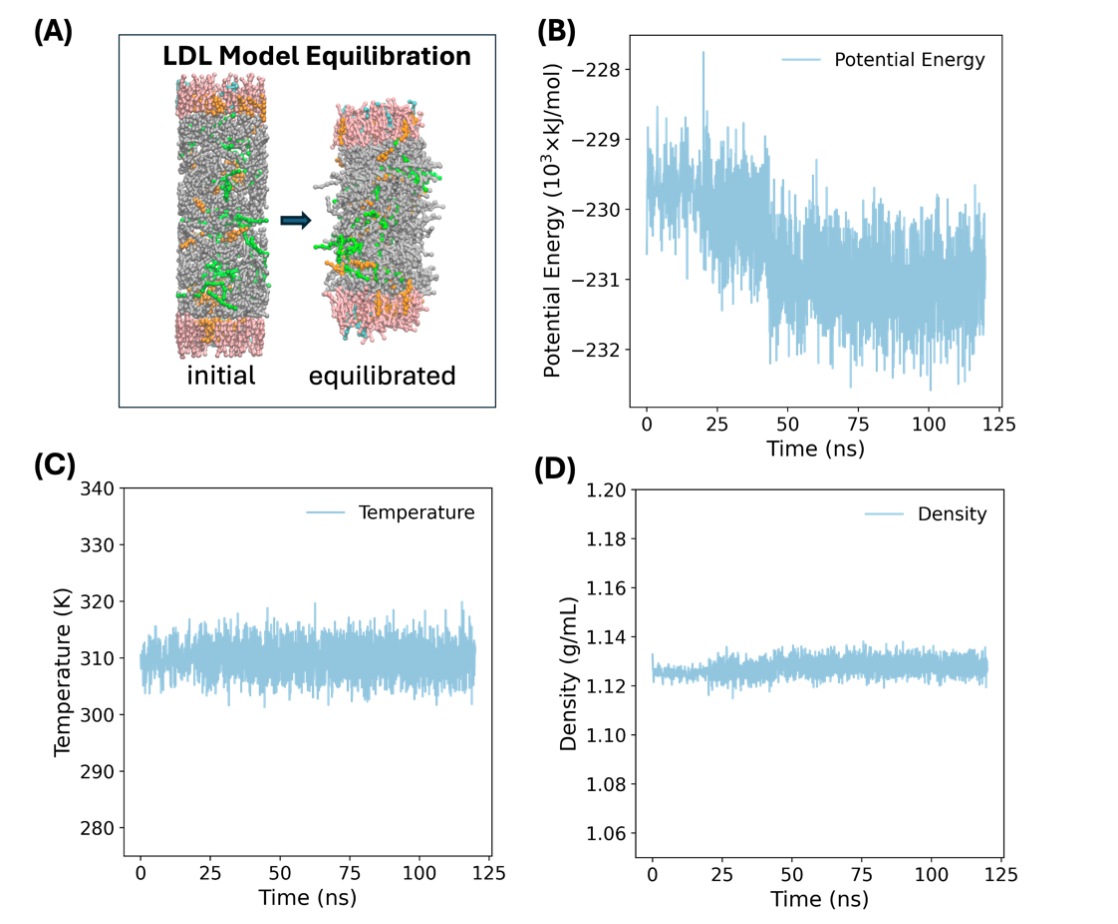
Figure 5: Illustration of the equilibration of the LDL model. (A) The snapshots of initial configuration and final equilibrated configuration of the LDL model. The phospholipid, POPC and LysoPC are shown in pink and cyan, respectively. The green, orange, gray color represent the TOG, cholesterol, and CHYO molecules. The convergence profiles of (B) potential energy, (C) temperature, and (D) density along the simulation time.
We further examine the distribution of molecules in the model since this is another important metric for evaluating the model. Figure 6 shows the partial density profiles of all components along the Z axis. The equilibrated snapshot is replicated and imposed for better visualization. From the partial density distribution, no water infiltration is observed into the LDL core because of the hydrophobic-hydrophilic repulsive interactions. Moreover, the partial overlapping of POPC and LysoPC heads with water phase, indicating that the membrane is well solvated by the water. In addition, the esters and cholesterols are well-distributed inside the LDL model, exhibiting a symmetric distribution in accordance with a number of bilayer models. All these observations are consistent with previous MD simulation on similar systems and suggest that the LDL model is well equilibrated, preserving the macroscopic properties and molecular details as in a real LDL droplet, and thus is suitable for subsequent modeling with drug models.
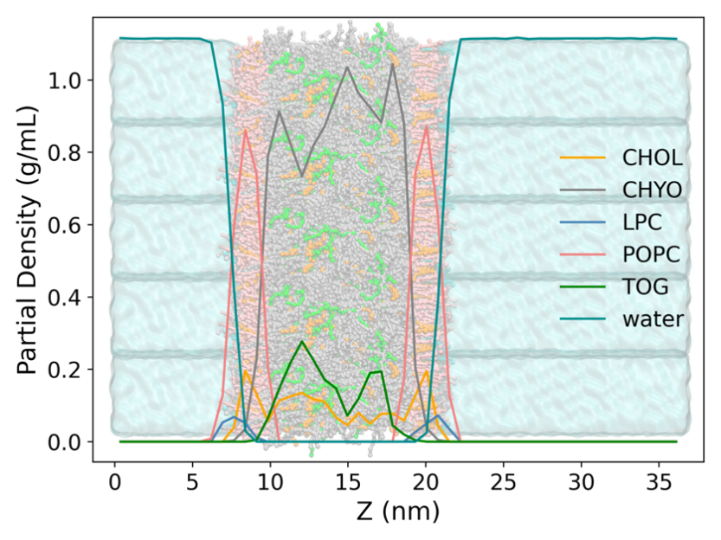
Figure 6: The partial density profiles of the LDL components. The LDL is shown in bonds and beads while the water is shown as a cyan bulk phase. The phospholipids are shown in pink and cyan, respectively. The green, orange, gray, pink and cyan colors represent the TOG, cholesterol, CHYO, POPC and LysoPC, respectively.
3.2. The Partition of Drug Molecules into LDL
With the established LDL model, we then attempt to model its interaction with the drugs. Two model drugs were chosen, including cladribine and dacarbazine. Cladribine is a purine antimetabolite used to treat multiple sclerosis (MS) and hairy cell leukemia [51]. dacarbazine is an alkylating agents used to treat melanoma, soft tissue sarcoma and Hodgkin lymphoma [52]. All these types of cancers have been reported to exhibit enhanced LDLR expression, and therefore the selected two drugs are well-suited for the study [31].
Cladribine and dacarbazine are both hydrophilic molecules and we expect that squalene conjugation can increase its interaction with LDL. One important consideration during model construction was that we made sure the linkage between the squalene to the drug is reversible under biological conditions and makes chemical sense. As a consequence, the ester linkage is used due to their propensity to hydrolysis in cells (e.g., lysosomes) and feasible synthesizability in lab. The chemical structures of the free drugs (CLA, DAC) and their squalene conjugates (SQ_CLA, SQ_DAC) are shown in Figure 3. Imposed on the chemical structures are the Martini beads used to model the drug molecules.
Subsequently, we inserted the drug molecules into the water phase as the initial structure and generated four intendent systems, with each containing a drug molecule. In principle, we can run a long conventional MD simulation and observe the partition of the drug molecules into LDL and use that for subsequent free energy calculations. However, in order to observe such transition would require microseconds or milliseconds simulations, which would pose a great challenge for us. Thus, we opted for steered MD simulation to gradually pull the molecule into the interior of LDL model along the Z axis using an external harmonic potential. The distance between the center of mass (COM) of the drug and LDL were monitored. Since this process is out-of-equilibrium, the pulling force and speed were carefully tuned to allow smooth transition while minimize the perturbation to the whole system. As shown in Figure 7A, a smooth decrease in the COM distance is observed implying that the pulling force was suitable, and the pulling process was slow enough to not to significantly perturb the equilibrium of the system. The representative configurations are also shown in Figure 7B to depict the partition process.
Although we obtained a series of configurations along the partition path, the process of drug entering the LDL is not necessarily energetically favored due to the existence of the artificial potential. We emphasize here that the major purpose of SMD simulations is to acquire the initial configuration (where the drug is within the LDL core) for the free energy calculations. The pulling process can never be regarded as the real partition process.
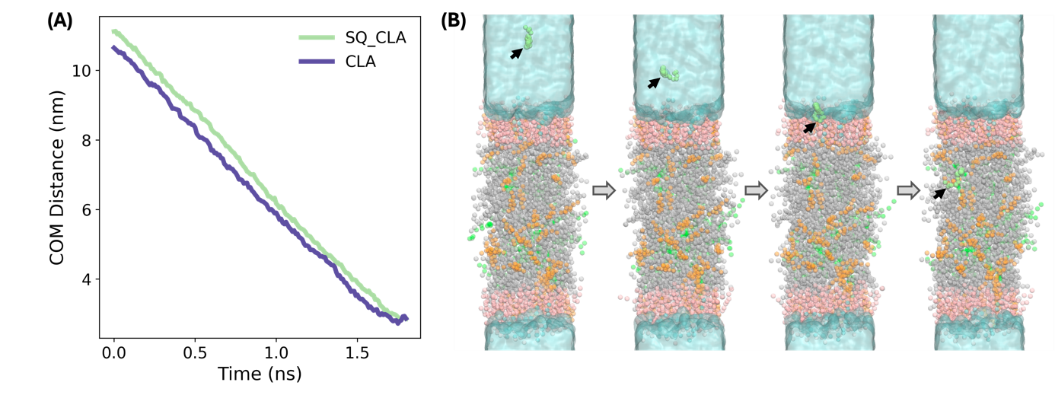
Figure 7: The SMD simulation for pulling the drug molecules into with CLA and SQ_CLA as an example. (A) The COM distance between CLA and SQ_CLA along the Z axis. (B) The representative snapshots from the SMD simulation to depict the process of drug partition into the LDL core. The green, orange, gray, pink and cyan colors represent the TOG, cholesterol, CHYO, POPC and LysoPC, respectively. The drug SQ_CLA is shown in lime green.
3.3. Squalene Conjugation Enhances the Drug Partition into LDL
Having acquired the configurations along the partition process, an additional step to calculate the free energies associated to the partition process is required. Thanks to the fact that free energy is a state function and is independent of the reaction path, we then avoided the use of umbrella sampling that enhances sampling along the whole pulling path, and opted for an alchemical transfer method that only determines the free energy difference between the predefined two states. The details of the implementation of the method can be found in the Methods section.
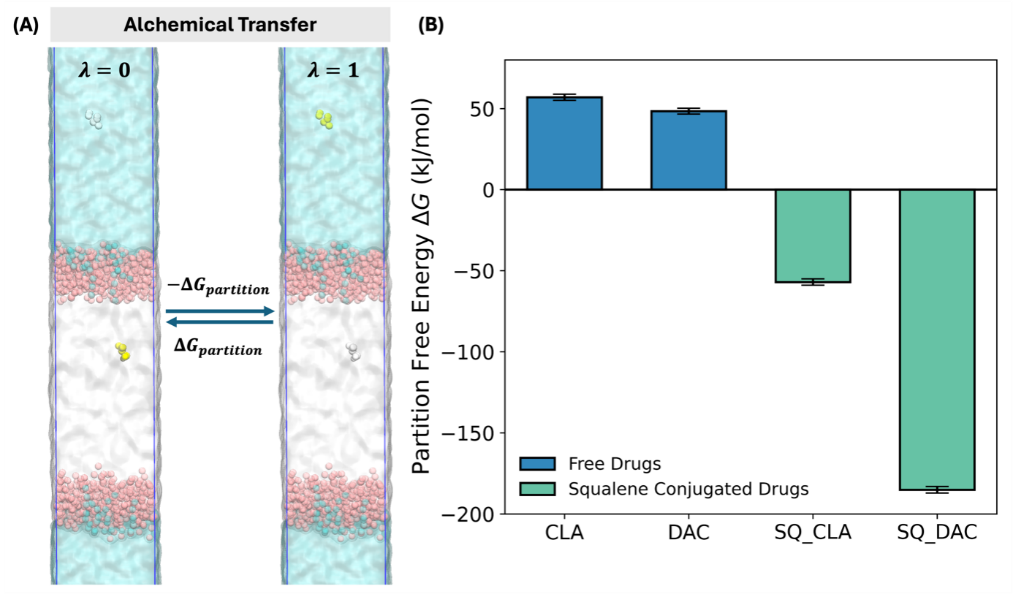
Figure 8: Illustration and results of the free energy calculation. (A) The schematic showing the alchemical transfer method. The interacting drug is shown in yellow while the non-interacting drug is shown in gray. The green, orange, gray, pink and cyan colors represent the TOG, cholesterol, CHYO, POPC and LysoPC, respectively. (B) Bar plot showing the partition free energy of the free drugs and their squalene conjugates.
As shown in Figure 8A, the two states are defined and controlled by the parameter λ. In state 0, the drug within the LDL is interacting with its environment, contributing to the potential energy of the system. In contrast, the drug in the water phase is non-interacting and thus does not interfere with the rest of the system. In state 1, interacting state is reversed with drug in the water phase interacting while that in the LDL not interacting. By slowing altering the λ value, we construct a series of windows that connect the two state. The free energy can thus be calculated using free energy perturbation theory.
The calculated free energies for all have converged within the simulation time (Figure 4) and are summarized in Figure 8B. It is obvious that due to the hydrophilic nature the hydrophilic free drugs show positive free energies of 56.92 kJ/mol and 48.37 kJ/mol for CLA and DAC, respectively. The positive free energy barrier suggests that the free drugs are not probable to enter the LDL interior and exploit the targeting ability of LDL particles. In contrast, the squalene conjugated drugs exhibit reversed free energies of -57.03 kJ/mol and -185.13 kJ/mol for SQ_CLA and SQ_DAC, respectively, leading to energy decreases of 113.95 kJ/mol and 233.50 kJ/mol. This huge energy benefit strongly indicate that the squalene conjugated drugs can bind to the LDL particle and progress to the interior for form a stable adduct. In such cases, the drug can enrich in the LDL during circulation in biological medium, which can subsequently accumulate in the cancer cells.
4. Conclusions and Discussions
Developing cancer targeting drugs, especially for hydrophilic chemodrugs, has been a long-standing research question as it possesses the benefits of improving the pharmacodynamics of drugs, enhancing treatment outcomes while reducing side effects(29, 53-56). In this contribution, we propose a novel approach by squalene conjugation to endow the drugs with the ability to exploit LDL particles to target cancer cells. Our MD simulation results show that the squalene conjugation to hydrophilic chemodrugs (CLA and DAC) is able to reverse and significantly reduce the free energy of the drug entering the LDL particle. During circulation in body, the squalene-drug conjugates may bind to and enrich in LDL particles first. Then, due to the enhanced expression of LDLR on certain cancer cells, the LDL particles will be scavenged and accumulate in tumor sites, along with the drugs within. In this way, the drug will also target to and function in cancer cells, and the elevated local drug concentration is expected to improve the cancer treatment effects and reduce its harm to healthy cells.
Since our project relies on computational studies, it has its own limitations that we aim to address in future. First, the CG model is an approximation for the interaction between molecules to achieve high computational efficiency, and the CG models for the drugs are not extensively parameterized under the assumption that current model can capture general trends [36]. Therefore, future study can focus on whether the results we obtained is consistent with all-atom simulations and gain more detailed chemical information. Secondly, the biological medium if a complex environment consisting of proteins, ions, and lipids, while modeling such is impossible in computers at least in the capability of our computation resources. Simplification of the computer model has to neglect certain factors, which may also affect the partition free energy and the kinetics. Last but not least, a gap generally exists between computational results an experimental observations, and thus experimental testing will be the next major step to prove the targeting concept.
Overall, we propose a new cancer targeting approach that a squalene conjugation can aid the drug in hitchhiking on endogenous LDL particles to target to cancer cells. We envision that such strategy can be generalized to a wide range of different chemodrugs, with the potential to improve cancer therapeutic outcomes, reducing side effects and eventually contributing to public health.
Acknowledgements
This thesis is the culmination of a long journey that I have only been able to make as a result of the dedicated support I have received from so many people along the way.
The deepest gratitude goes to my teacher Mr. Liu for his continuous guidance throughout my research. I first found my interest in finding cancer treatment methods during the communication with him. I found that the current methods have various side effects, and I wondered if I could do some research on it. I shared this idea with him and finally set our research topic to be the drugs for enhanced targeting for cancer therapy. During the whole researching process, he spent a lot of time communicating with me for the researching methods, tools, and techniques that I had to use or learn. We used the LDL model to simulate the particles and did the equilibration. During the process, Mr. Liu taught me some basic knowledge about the model to assist me get the data needed. I also modeled the drug molecules. For computing the partition free energy using an alchemical transfer approach, we used the steered MD simulations. Mr. Liu also gave me much support during the simulation. Then we employed an alchemical transfer protocol. Briefly, we copied the drug in the final configuration from the SMD simulation to the water phase and did free energy calculation. During the process of composing the thesis, Mr. Liu provided me with professional instructions on the language, word usage and some other points.
While doing the simulations, there were also some obstacles. We used the terminal on the MacBook to download some files of models for simulation. However, the compilation of the instructions always turns out to have errors. Mr. Liu always helped me search for solutions or other resources. Sometimes we could even try many times for one file. To get a proper setting of the simulation environment, we had to try many different conditions such as temperature to find the best one.
Also, I appreciate my school Shanghai Starriver Bilingual School for its hardware support. It provided me with a stable simulation platform online to do my research more efficiently and allowed me to keep all the data on the platform.
Moreover, I am extremely grateful to my parents for their support, motive, patience, love and care during my research. Without their help, this thesis would not have been accomplished. I also want to thank my classmates and friends who always encouraged me and tried their best to help me whenever I needed information or experience. My final appreciation go to all people that have directly or indirectly supported me during the process. Everyone I’ve mentioned has helped me to improve, grow, and become a better person. They also made me more curious about science. Sincere wishes to all of you.
References
[1]. H. Sung et al., Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 71, 209-249 (2021).
[2]. Erratum to "Cancer statistics, 2024". CA Cancer J Clin 74, 203 (2024).
[3]. R. L. Siegel, K. D. Miller, N. S. Wagle, A. Jemal, Cancer statistics, 2023. CA Cancer J Clin 73, 17-48 (2023).
[4]. R. L. Siegel, K. D. Miller, H. E. Fuchs, A. Jemal, Cancer statistics, 2022. CA Cancer J Clin 72, 7-33 (2022).
[5]. D. Hanahan, Hallmarks of Cancer: New Dimensions. Cancer Discov 12, 31-46 (2022).
[6]. L. Toschi, G. Finocchiaro, S. Bartolini, V. Gioia, F. Cappuzzo, Role of gemcitabine in cancer therapy. Future Oncology 1, 7-17 (2005).
[7]. S. L. Koolen et al., Phase I study of Oral gemcitabine prodrug (LY2334737) alone and in combination with erlotinib in patients with advanced solid tumors. Clin Cancer Res 17, 6071-6082 (2011).
[8]. J. Ciccolini, C. Serdjebi, G. J. Peters, E. Giovannetti, Pharmacokinetics and pharmacogenetics of Gemcitabine as a mainstay in adult and pediatric oncology: an EORTC-PAMM perspective. Cancer Chemotherapy and Pharmacology 78, 1-12 (2016).
[9]. M. Mehrmohamadi, S. H. Jeong, J. W. Locasale, Molecular features that predict the response to antimetabolite chemotherapies. Cancer & Metabolism 5, 8 (2017).
[10]. H. E. Barker, J. T. Paget, A. A. Khan, K. J. Harrington, The tumour microenvironment after radiotherapy: mechanisms of resistance and recurrence. Nat Rev Cancer 15, 409-425 (2015).
[11]. K. Wang, J. E. Tepper, Radiation therapy-associated toxicity: Etiology, management, and prevention. CA Cancer J Clin 71, 437-454 (2021).
[12]. J. Le Naour, L. Zitvogel, L. Galluzzi, E. Vacchelli, G. Kroemer, Trial watch: STING agonists in cancer therapy. Oncoimmunology 9, (2020).
[13]. K. Kato et al., Structural insights into cGAMP degradation by Ecto-nucleotide pyrophosphatase phosphodiesterase 1. Nature Communications 9, (2018).
[14]. S. Crunkhorn, Strengthening the sting of immunotherapy. Nat Rev Drug Discov 19, 669-669 (2020).
[15]. Y.-Y. Wang, Y.-C. Liu, H. Sun, D.-S. Guo, Type I photodynamic therapy by organic–inorganic hybrid materials: From strategies to applications. Coord Chem Rev 395, 46-62 (2019).
[16]. Y.-Y. Huang et al., Stable Synthetic Bacteriochlorins for Photodynamic Therapy: Role of Dicyano Peripheral Groups, Central Metal Substitution (2H, Zn, Pd), and Cremophor EL Delivery. ChemMedChem 7, 2155-2167 (2012).
[17]. G. Gunaydin, M. E. Gedik, S. Ayan, Photodynamic Therapy-Current Limitations and Novel Approaches. Front Chem 9, 691697 (2021).
[18]. E. Dalla Pozza et al., Targeting gemcitabine containing liposomes to CD44 expressing pancreatic adenocarcinoma cells causes an increase in the antitumoral activity. Biochim Biophys Acta 1828, 1396-1404 (2013).
[19]. C. R. Patra et al., Targeted Delivery of Gemcitabine to Pancreatic Adenocarcinoma Using Cetuximab as a Targeting Agent. Cancer Research 68, 1970-1978 (2008).
[20]. E. D. Brooks, J. Y. Chang, Time to abandon single-site irradiation for inducing abscopal effects. Nature Reviews Clinical Oncology 16, 123-135 (2019).
[21]. B. Venugopal et al., A first-in-human phase I and pharmacokinetic study of CP-4126 (CO-101), a nucleoside analogue, in patients with advanced solid tumours. Cancer Chemother Pharmacol 76, 785-792 (2015).
[22]. A. H. Calvert et al., Carboplatin dosage: prospective evaluation of a simple formula based on renal function. J Clin Oncol 41, 4453-4454 (2023).
[23]. B. T. Oronsky, T. Reid, S. J. Knox, J. J. Scicinski, The scarlet letter of alkylation: a mini review of selective alkylating agents. Transl Oncol 5, 226-229 (2012).
[24]. Y. Pommier, Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer 6, 789-802 (2006).
[25]. Y. Pommier, M. Cushman, J. H. Doroshow, Novel clinical indenoisoquinoline topoisomerase I inhibitors: a twist around the camptothecins. Oncotarget 9, 37286-37288 (2018).
[26]. D. Fan et al., Nanomedicine in cancer therapy. Signal Transduction and Targeted Therapy 8, 293 (2023).
[27]. Z. Li, J. Zou, X. Chen, In Response to Precision Medicine: Current Subcellular Targeting Strategies for Cancer Therapy. Adv Mater 35, e2209529 (2023).
[28]. N. Niu et al., A cell membrane-targeting AIE photosensitizer as a necroptosis inducer for boosting cancer theranostics. Chem Sci 13, 5929-5937 (2022).
[29]. O. C. Ubah, H. M. Wallace, Cancer therapy: Targeting mitochondria and other sub-cellular organelles. Curr Pharm Des 20, 201-222 (2014).
[30]. L. Maran, A. Hamid, S. B. S. Hamid, Lipoproteins as Markers for Monitoring Cancer Progression. Journal of Lipids 2021, 8180424 (2021).
[31]. C. F. Deng et al., Involvement of LDL and ox-LDL in Cancer Development and Its Therapeutical Potential. Front Oncol 12, 803473 (2022).
[32]. X. Guan et al., Emerging roles of low-density lipoprotein in the development and treatment of breast cancer. Lipids in Health and Disease 18, 137 (2019).
[33]. I. Rajman, P. I. Eacho, P. J. Chowienczyk, J. M. Ritter, LDL particle size: an important drug target? Br J Clin Pharmacol 48, 125-133 (1999).
[34]. S.-K. Kim, F. Karadeniz, in Advances in Food and Nutrition Research, S.-K. Kim, Ed. (Academic Press, 2012), vol. 65, pp. 223-233.
[35]. T. Murtola et al., Low density lipoprotein: structure, dynamics, and interactions of apoB-100 with lipids. Soft Matter 7, 8135-8141 (2011).
[36]. S. J. Marrink, H. J. Risselada, S. Yefimov, D. P. Tieleman, A. H. de Vries, The MARTINI Force Field: Coarse Grained Model for Biomolecular Simulations. The Journal of Physical Chemistry B 111, 7812-7824 (2007).
[37]. L. Martinez, R. Andrade, E. G. Birgin, J. M. Martinez, PACKMOL: a package for building initial configurations for molecular dynamics simulations. J Comput Chem 30, 2157-2164 (2009).
[38]. M. J. Abraham et al., GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2, 19-25 (2015).
[39]. O. Farago, Langevin thermostat for robust configurational and kinetic sampling. Physica A: Statistical Mechanics and its Applications 534, 122210 (2019).
[40]. Y. Lin, D. Pan, J. Li, L. Zhang, X. Shao, Application of Berendsen barostat in dissipative particle dynamics for nonequilibrium dynamic simulation. The Journal of Chemical Physics 146, (2017).
[41]. M. Bernetti, G. Bussi, Pressure control using stochastic cell rescaling. J Chem Phys 153, 114107 (2020).
[42]. W. Humphrey, A. Dalke, K. Schulten, VMD: visual molecular dynamics. J Mol Graph 14, 33-38, 27-38 (1996).
[43]. M. Floeth et al., Low-density lipoprotein receptor (LDLR) is an independent adverse prognostic factor in acute myeloid leukaemia. British Journal of Haematology 192, 494-503 (2021).
[44]. T. D. Potter, E. L. Barrett, M. A. Miller, Automated Coarse-Grained Mapping Algorithm for the Martini Force Field and Benchmarks for Membrane-Water Partitioning. J Chem Theory Comput 17, 5777-5791 (2021).
[45]. F. S. Zariquiey, A. Perez, M. Majewski, E. Gallicchio, G. Fabritiis, Validation of the Alchemical Transfer Method for the Estimation of Relative Binding Affinities of Molecular Series. ArXiv, (2023).
[46]. J. G. Kirkwood, Statistical Mechanics of Fluid Mixtures. The Journal of Chemical Physics 3, 300-313 (1935).
[47]. C. H. Bennett, Efficient estimation of free energy differences from Monte Carlo data. Journal of Computational Physics 22, 245-268 (1976).
[48]. M. R. Shirts, J. D. Chodera, Statistically optimal analysis of samples from multiple equilibrium states. J Chem Phys 129, 124105 (2008).
[49]. J. D. Chodera, A Simple Method for Automated Equilibration Detection in Molecular Simulations. J Chem Theory Comput 12, 1799-1805 (2016).
[50]. S. O. Yesylevskyy, C. Ramseyer, M. Savenko, S. Mura, P. Couvreur, Low-Density Lipoproteins and Human Serum Albumin as Carriers of Squalenoylated Drugs: Insights from Molecular Simulations. Molecular Pharmaceutics 15, 585-591 (2018).
[51]. F. Nabizadeh et al., Safety and efficacy of cladribine in multiple sclerosis: a systematic review and meta-analysis. Neurological Sciences 44, 3045-3057 (2023).
[52]. A. M. M. Eggermont, J. M. Kirkwood, Re-evaluating the role of dacarbazine in metastatic melanoma: what have we learned in 30 years? European Journal of Cancer 40, 1825-1836 (2004).
[53]. K. Ni, G. Lan, Y. Song, Z. Hao, W. Lin, Biomimetic nanoscale metal-organic framework harnesses hypoxia for effective cancer radiotherapy and immunotherapy. Chem Sci 11, 7641-7653 (2020).
[54]. U. Anand et al., Cancer chemotherapy and beyond: Current status, drug candidates, associated risks and progress in targeted therapeutics. Genes Dis 10, 1367-1401 (2023).
[55]. J. D. Twomey, B. L. Zhang, Cancer Immunotherapy Update: FDA-Approved Checkpoint Inhibitors and Companion Diagnostics. Aaps J 23, (2021).
[56]. J. Shi, P. W. Kantoff, R. Wooster, O. C. Farokhzad, Cancer nanomedicine: progress, challenges and opportunities. Nat Rev Cancer 17, 20-37 (2017).
Cite this article
Yuan,Z. (2025). Squalene-Conjugated Drugs Exploit Endogenous Low-Density Lipoproteins for Enhanced Targeting for Cancer Therapy. Theoretical and Natural Science,68,57-69.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 3rd International Conference on Modern Medicine and Global Health
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. H. Sung et al., Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 71, 209-249 (2021).
[2]. Erratum to "Cancer statistics, 2024". CA Cancer J Clin 74, 203 (2024).
[3]. R. L. Siegel, K. D. Miller, N. S. Wagle, A. Jemal, Cancer statistics, 2023. CA Cancer J Clin 73, 17-48 (2023).
[4]. R. L. Siegel, K. D. Miller, H. E. Fuchs, A. Jemal, Cancer statistics, 2022. CA Cancer J Clin 72, 7-33 (2022).
[5]. D. Hanahan, Hallmarks of Cancer: New Dimensions. Cancer Discov 12, 31-46 (2022).
[6]. L. Toschi, G. Finocchiaro, S. Bartolini, V. Gioia, F. Cappuzzo, Role of gemcitabine in cancer therapy. Future Oncology 1, 7-17 (2005).
[7]. S. L. Koolen et al., Phase I study of Oral gemcitabine prodrug (LY2334737) alone and in combination with erlotinib in patients with advanced solid tumors. Clin Cancer Res 17, 6071-6082 (2011).
[8]. J. Ciccolini, C. Serdjebi, G. J. Peters, E. Giovannetti, Pharmacokinetics and pharmacogenetics of Gemcitabine as a mainstay in adult and pediatric oncology: an EORTC-PAMM perspective. Cancer Chemotherapy and Pharmacology 78, 1-12 (2016).
[9]. M. Mehrmohamadi, S. H. Jeong, J. W. Locasale, Molecular features that predict the response to antimetabolite chemotherapies. Cancer & Metabolism 5, 8 (2017).
[10]. H. E. Barker, J. T. Paget, A. A. Khan, K. J. Harrington, The tumour microenvironment after radiotherapy: mechanisms of resistance and recurrence. Nat Rev Cancer 15, 409-425 (2015).
[11]. K. Wang, J. E. Tepper, Radiation therapy-associated toxicity: Etiology, management, and prevention. CA Cancer J Clin 71, 437-454 (2021).
[12]. J. Le Naour, L. Zitvogel, L. Galluzzi, E. Vacchelli, G. Kroemer, Trial watch: STING agonists in cancer therapy. Oncoimmunology 9, (2020).
[13]. K. Kato et al., Structural insights into cGAMP degradation by Ecto-nucleotide pyrophosphatase phosphodiesterase 1. Nature Communications 9, (2018).
[14]. S. Crunkhorn, Strengthening the sting of immunotherapy. Nat Rev Drug Discov 19, 669-669 (2020).
[15]. Y.-Y. Wang, Y.-C. Liu, H. Sun, D.-S. Guo, Type I photodynamic therapy by organic–inorganic hybrid materials: From strategies to applications. Coord Chem Rev 395, 46-62 (2019).
[16]. Y.-Y. Huang et al., Stable Synthetic Bacteriochlorins for Photodynamic Therapy: Role of Dicyano Peripheral Groups, Central Metal Substitution (2H, Zn, Pd), and Cremophor EL Delivery. ChemMedChem 7, 2155-2167 (2012).
[17]. G. Gunaydin, M. E. Gedik, S. Ayan, Photodynamic Therapy-Current Limitations and Novel Approaches. Front Chem 9, 691697 (2021).
[18]. E. Dalla Pozza et al., Targeting gemcitabine containing liposomes to CD44 expressing pancreatic adenocarcinoma cells causes an increase in the antitumoral activity. Biochim Biophys Acta 1828, 1396-1404 (2013).
[19]. C. R. Patra et al., Targeted Delivery of Gemcitabine to Pancreatic Adenocarcinoma Using Cetuximab as a Targeting Agent. Cancer Research 68, 1970-1978 (2008).
[20]. E. D. Brooks, J. Y. Chang, Time to abandon single-site irradiation for inducing abscopal effects. Nature Reviews Clinical Oncology 16, 123-135 (2019).
[21]. B. Venugopal et al., A first-in-human phase I and pharmacokinetic study of CP-4126 (CO-101), a nucleoside analogue, in patients with advanced solid tumours. Cancer Chemother Pharmacol 76, 785-792 (2015).
[22]. A. H. Calvert et al., Carboplatin dosage: prospective evaluation of a simple formula based on renal function. J Clin Oncol 41, 4453-4454 (2023).
[23]. B. T. Oronsky, T. Reid, S. J. Knox, J. J. Scicinski, The scarlet letter of alkylation: a mini review of selective alkylating agents. Transl Oncol 5, 226-229 (2012).
[24]. Y. Pommier, Topoisomerase I inhibitors: camptothecins and beyond. Nat Rev Cancer 6, 789-802 (2006).
[25]. Y. Pommier, M. Cushman, J. H. Doroshow, Novel clinical indenoisoquinoline topoisomerase I inhibitors: a twist around the camptothecins. Oncotarget 9, 37286-37288 (2018).
[26]. D. Fan et al., Nanomedicine in cancer therapy. Signal Transduction and Targeted Therapy 8, 293 (2023).
[27]. Z. Li, J. Zou, X. Chen, In Response to Precision Medicine: Current Subcellular Targeting Strategies for Cancer Therapy. Adv Mater 35, e2209529 (2023).
[28]. N. Niu et al., A cell membrane-targeting AIE photosensitizer as a necroptosis inducer for boosting cancer theranostics. Chem Sci 13, 5929-5937 (2022).
[29]. O. C. Ubah, H. M. Wallace, Cancer therapy: Targeting mitochondria and other sub-cellular organelles. Curr Pharm Des 20, 201-222 (2014).
[30]. L. Maran, A. Hamid, S. B. S. Hamid, Lipoproteins as Markers for Monitoring Cancer Progression. Journal of Lipids 2021, 8180424 (2021).
[31]. C. F. Deng et al., Involvement of LDL and ox-LDL in Cancer Development and Its Therapeutical Potential. Front Oncol 12, 803473 (2022).
[32]. X. Guan et al., Emerging roles of low-density lipoprotein in the development and treatment of breast cancer. Lipids in Health and Disease 18, 137 (2019).
[33]. I. Rajman, P. I. Eacho, P. J. Chowienczyk, J. M. Ritter, LDL particle size: an important drug target? Br J Clin Pharmacol 48, 125-133 (1999).
[34]. S.-K. Kim, F. Karadeniz, in Advances in Food and Nutrition Research, S.-K. Kim, Ed. (Academic Press, 2012), vol. 65, pp. 223-233.
[35]. T. Murtola et al., Low density lipoprotein: structure, dynamics, and interactions of apoB-100 with lipids. Soft Matter 7, 8135-8141 (2011).
[36]. S. J. Marrink, H. J. Risselada, S. Yefimov, D. P. Tieleman, A. H. de Vries, The MARTINI Force Field: Coarse Grained Model for Biomolecular Simulations. The Journal of Physical Chemistry B 111, 7812-7824 (2007).
[37]. L. Martinez, R. Andrade, E. G. Birgin, J. M. Martinez, PACKMOL: a package for building initial configurations for molecular dynamics simulations. J Comput Chem 30, 2157-2164 (2009).
[38]. M. J. Abraham et al., GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2, 19-25 (2015).
[39]. O. Farago, Langevin thermostat for robust configurational and kinetic sampling. Physica A: Statistical Mechanics and its Applications 534, 122210 (2019).
[40]. Y. Lin, D. Pan, J. Li, L. Zhang, X. Shao, Application of Berendsen barostat in dissipative particle dynamics for nonequilibrium dynamic simulation. The Journal of Chemical Physics 146, (2017).
[41]. M. Bernetti, G. Bussi, Pressure control using stochastic cell rescaling. J Chem Phys 153, 114107 (2020).
[42]. W. Humphrey, A. Dalke, K. Schulten, VMD: visual molecular dynamics. J Mol Graph 14, 33-38, 27-38 (1996).
[43]. M. Floeth et al., Low-density lipoprotein receptor (LDLR) is an independent adverse prognostic factor in acute myeloid leukaemia. British Journal of Haematology 192, 494-503 (2021).
[44]. T. D. Potter, E. L. Barrett, M. A. Miller, Automated Coarse-Grained Mapping Algorithm for the Martini Force Field and Benchmarks for Membrane-Water Partitioning. J Chem Theory Comput 17, 5777-5791 (2021).
[45]. F. S. Zariquiey, A. Perez, M. Majewski, E. Gallicchio, G. Fabritiis, Validation of the Alchemical Transfer Method for the Estimation of Relative Binding Affinities of Molecular Series. ArXiv, (2023).
[46]. J. G. Kirkwood, Statistical Mechanics of Fluid Mixtures. The Journal of Chemical Physics 3, 300-313 (1935).
[47]. C. H. Bennett, Efficient estimation of free energy differences from Monte Carlo data. Journal of Computational Physics 22, 245-268 (1976).
[48]. M. R. Shirts, J. D. Chodera, Statistically optimal analysis of samples from multiple equilibrium states. J Chem Phys 129, 124105 (2008).
[49]. J. D. Chodera, A Simple Method for Automated Equilibration Detection in Molecular Simulations. J Chem Theory Comput 12, 1799-1805 (2016).
[50]. S. O. Yesylevskyy, C. Ramseyer, M. Savenko, S. Mura, P. Couvreur, Low-Density Lipoproteins and Human Serum Albumin as Carriers of Squalenoylated Drugs: Insights from Molecular Simulations. Molecular Pharmaceutics 15, 585-591 (2018).
[51]. F. Nabizadeh et al., Safety and efficacy of cladribine in multiple sclerosis: a systematic review and meta-analysis. Neurological Sciences 44, 3045-3057 (2023).
[52]. A. M. M. Eggermont, J. M. Kirkwood, Re-evaluating the role of dacarbazine in metastatic melanoma: what have we learned in 30 years? European Journal of Cancer 40, 1825-1836 (2004).
[53]. K. Ni, G. Lan, Y. Song, Z. Hao, W. Lin, Biomimetic nanoscale metal-organic framework harnesses hypoxia for effective cancer radiotherapy and immunotherapy. Chem Sci 11, 7641-7653 (2020).
[54]. U. Anand et al., Cancer chemotherapy and beyond: Current status, drug candidates, associated risks and progress in targeted therapeutics. Genes Dis 10, 1367-1401 (2023).
[55]. J. D. Twomey, B. L. Zhang, Cancer Immunotherapy Update: FDA-Approved Checkpoint Inhibitors and Companion Diagnostics. Aaps J 23, (2021).
[56]. J. Shi, P. W. Kantoff, R. Wooster, O. C. Farokhzad, Cancer nanomedicine: progress, challenges and opportunities. Nat Rev Cancer 17, 20-37 (2017).









