1. Introduction
Meiosis is a specialized form of cell division that generates haploid gametes, essential for maintaining chromosomal stability across generations and promoting genetic diversity [1]. This process, unique to eukaryotes, involves genome-wide reorganization, including homolog recognition, pairing, synapsis via the synaptonemal complex (SC), and recombination. At least one crossover per homolog pair is required to ensure accurate chromosome segregation; failure to do so can result in aneuploidy, leading to infertility or developmental disorders such as Down syndrome. To safeguard this fidelity, most eukaryotes have evolved meiotic checkpoints that monitor homolog pairing and recombination during early prophase I.
Genetic model organisms have been indispensable for elucidating meiotic mechanisms. Unicellular systems such as yeast provide powerful genetic and biochemical toolkits but lack tissue complexity. In contrast, Drosophila and mouse offer physiological relevance yet are constrained by limited meiotic cell numbers, longer generation times, and challenges in live-cell imaging. Caenorhabditis elegans strikes a favorable balance between experimental tractability and biological relevance.
Adult C. elegans harbor a linear gonad containing thousands of nuclei arranged along a temporal–spatial meiotic gradient, enabling high-resolution in vivo imaging of meiosis within intact tissues. A short life cycle (3 days at 25 °C), high fecundity, and optical transparency facilitate large-scale genetic screens and dynamic analyses of chromosome behavior [2]. The species’ genetic toolkit includes genome-wide RNA interference (RNAi), efficient CRISPR/Cas9-based genome editing, and transgenic lines expressing fluorescently tagged proteins. These approaches have revealed conserved meiotic regulators such as msh-4, msh-5, and cosa-1, and enabled quantitative, live analyses of SC dynamics and chromosome motions.
Classical forward-genetic screens in C. elegans identified key meiotic mutant classes, including him (“high incidence of males”) and spe (“spermatogenesis defective”), underscoring the power of this system for dissecting chromosome segregation pathways. Collectively, the developmental context, cellular accessibility, and experimental versatility of C. elegans establish it as an ideal platform for resolving the molecular logic of meiosis, with current research spanning homolog pairing and recombination, chromosome segregation and spindle assembly, and checkpoint signaling—and poised to benefit from emerging technologies and comparative approaches.
2. Model advantages
2.1. Genomic and genetic advantages
2.1.1 Compact genome with high-quality annotation
C. elegans possesses a compact genome (about 100 Mb) distributed across six chromosomes (five autosomes and one X chromosome), encoding approximately 20,000 protein-coding genes. It was among the first multicellular organisms to be fully sequenced, and its gene models, annotations, and mutant libraries are highly curated. The genome features short intergenic regions, low repeat content, and predominantly single-copy genes, facilitating cloning, mapping, and whole-genome analyses. Notably, worm chromosomes are holocentric, with kinetochores distributed along their entire length. This architecture permits microtubule attachment to chromosomal fragments, potentially enhancing tolerance to chromosomal breaks and offering unique experimental advantages.
2.1.2 Genomic stability and low transposon activity
The standard laboratory strain N2 exhibits low transposon content (about 12–16%) and strong transposable element silencing mechanisms, including the 21U-RNA/piRNA pathway [3]. Consequently, most transposons (e.g., Tc1, Tc3) are either mutated or transcriptionally repressed. This contributes to an exceptionally low spontaneous mutation rate (about 10⁻⁹ per base per generation), enabling consistent genetic backgrounds across generations. Such stability supports reproducibility in genetic and meiotic studies by minimizing background variation.
2.1.3 Versatile genetic toolkit
C. elegans offers an advanced and scalable system for genetic manipulation. It was a pioneer in genome-wide RNA interference (RNAi), enabled by feeding worms with E. coli expressing double-stranded RNA targeting specific genes. The syncytial architecture of the adult germline enhances RNAi efficacy in meiotic cells, supporting rapid functional genomics. Large-scale RNAi screens have identified thousands of genes involved in germline and embryonic development.
Moreover, CRISPR/Cas9-based genome editing is highly efficient in worms. Microinjection of Cas9–sgRNA complexes into zygotes allows precise introduction of mutations, epitope tags, or conditional alleles at relatively low cost and high throughput [2]. Additional tools—such as balancer chromosomes, degron systems, and recombineering—enable fine-tuned genetic control. Established reporter lines (e.g., HIM-8::GFP) allow live visualization of meiotic events, including homolog pairing and synapsis.
Together, these features render C. elegans a genetically tractable system for dissecting gene function in meiosis with precision, scalability, and reproducibility.
2.2. Morphological and imaging advantages
The gonad of hermaphrodite C. elegans is linear and spatially organized, with germ cells arranged along a distal–proximal axis. Mitotic germline stem cells at the distal tip give rise to nuclei that sequentially progress through all meiotic prophase I stages within a single tissue [4]. This anatomical layout enables direct side-by-side analysis of meiotic transitions without the need for staging or cell isolation.
The worm’s optical transparency facilitates live imaging of meiotic processes using fluorescent reporters and confocal microscopy. Dynamic events such as SC assembly, chromosome movement, and spindle formation can be visualized in vivo at subcellular resolution.
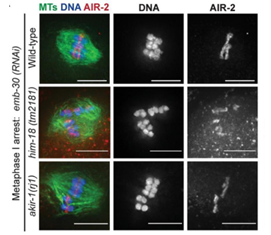
Moreover, meiotic proteins can be easily tagged or immunostained. All SC components (SYP-1–4), kinetochores (e.g., AIR-2), and cohesins have been visualized in fixed or live germlines. Super-resolution techniques like SIM and STED have revealed fine structures, such as microtubule channels within the ring complex, undetectable by conventional microscopy [5]. In addition, as shown in Figure 1, high-resolution imaging enables detailed analysis of chromosome alignment on the meiotic spindle in C. elegans, and allows quantitative assessment of subtle differences in the position and morphology of chromosomes on the spindle across different mutants (Figure 1).
Together, the gonad architecture and imaging accessibility make C. elegans uniquely suited for high-resolution analysis of meiotic structures and dynamics.
2.3. Reproductive and developmental advantages
2.3.1 Synchronized and continuous development
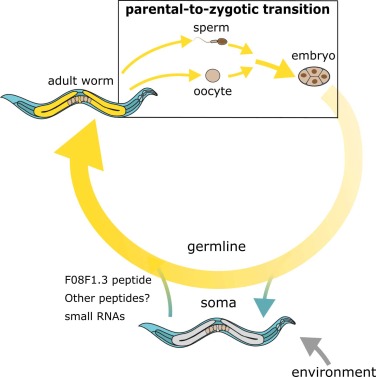
C. elegans hermaphrodites produce hundreds of oocytes in a spatially ordered gonad, where germ cells progress synchronously from mitotic proliferation to meiotic entry, homolog pairing, synapsis, and finally to fertilization. The gonad’s distal–proximal axis captures this temporal sequence: mitotic cells occupy the distal tip, followed by meiotic nuclei in prophase I, maturing oocytes at diplotene/diakinesis, and post-meiotic oocytes in the uterus. This arrangement enables researchers to visualize all major meiotic stages in a single animal without staging or dissection of multiple specimens. It facilitates direct comparisons across meiotic stages and simplifies mutant phenotyping, particularly for processes such as synaptonemal complex dynamics, crossover regulation, or the transition into embryogenesis. As shown in Figure 2, the complete life cycle of C. elegans clearly illustrates the transition from parent to zygote, providing a direct visualization of germline continuity as well as the interactions between environment and individual, and between somatic and germ cells (Figure 2).
2.3.2 Classical genetic screens in C. elegans have identified many meiosis-specific mutants
Forward genetic screens have revealed a wide array of mutants affecting meiotic processes. The him (high incidence of males) class identifies genes involved in X chromosome segregation, while spe (spermatogenesis-defective) mutants uncover defects in sperm development. These studies underscore the power of the worm model in systematically uncovering genes that regulate meiotic progression and germ cell differentiation.
2.3.3 Self-fertilization with outcrossing
C. elegans reproduces primarily by self-fertilization, with each hermaphrodite producing 300 sperm during late larval stages and subsequently switching to oocyte production [6]. Selfing allows rapid homozygosity of recessive alleles and simplifies maintenance of mutant lines. However, males (XO) arise spontaneously at low frequency and can be used for outcrossing, enabling recombination, linkage analysis, and construction of double or triple mutants. Mating also increases brood size, enhancing experimental throughput. This dual-mode reproduction balances genetic stability with flexibility for complex genetic manipulation.
2.3.4 Dual spermatogenesis and oogenesis
Hermaphrodites undergo both spermatogenesis and oogenesis, providing access to male and female meiotic programs within a single individual. Although spermatogenesis is brief and yields fewer cells, male meiosis can be expanded using him-8, fog-2, or other masculinizing mutations to enrich for spermatocytes. Notably, worm sperm are amoeboid and undergo highly asymmetric divisions, distinct from canonical flagellated sperm. In contrast, oocyte meiosis involves acentrosomal spindle assembly, resembling mechanisms in mammalian oocytes. This duality allows comparative studies of sex-specific features of meiosis under genetically identical backgrounds.
Taken together, C. elegans in its genomic/genetic makeup, gonad structure, and reproductive strategy offers a powerful model system for meiosis research. Its unique combination of synchronized development, dual gametogenesis, and experimental tractability enables detailed analysis of both shared and sex-specific mechanisms governing meiotic progression.
3. Advances in meiotic mechanism research
3.1. Chromosome pairing and recombination mechanisms
3.1.1 Homolog recognition and pairing
In early prophase I, homologous chromosomes in Caenorhabditis elegans recognize each other through a unique pairing center (PC) mechanism. Each chromosome contains a PC near one end that binds specific zinc-finger proteins: HIM-8 for the X chromosome, and ZIM-1/2/3 for autosomes I–V [1]. These proteins tether chromosomes to the nuclear envelope and recruit factors such as PLK-2, promoting dynamic chromosomal movements. Interaction between homologous PCs is facilitated by SUN/KASH proteins, enhancing the likelihood of homology-based encounters. Once contact is established, chromosomes align along their lengths, initiating synapsis. Remarkably, this PC-driven alignment can occur even in the absence of Spo11-induced double-strand breaks (DSBs), although crossovers do not form. Conversely, disruption of PCs (e.g., him-8 or zim mutants) impairs pairing despite the presence of DSBs, underscoring the PC’s primary role in initiating homolog pairing.
3.1.2 Synaptonemal complex assembly and crossover control
Following alignment, the synaptonemal complex (SC) assembles between homologous axes. The SC comprises lateral element proteins (HIM-3, HTP-1/2/3) and central element proteins (SYP-1 to SYP-4). Its assembly stabilizes pairing and supports DSB formation and repair. C. elegans typically generates one crossover (CO) per homolog pair, enforced by strong interference and assurance mechanisms. This pattern results from limiting DSBs and directing repair toward a single CO via the MutSγ complex (MSH-4/MSH-5) and the cyclin-like protein COSA-1, which marks the designated CO site. Non-designated DSBs are repaired via noncrossover pathways. Cells lacking at least one CO per homolog trigger meiotic checkpoints and undergo apoptosis.
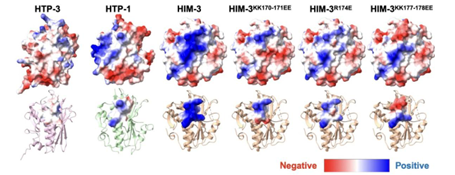
Recent studies have significantly advanced our understanding of the intrinsic properties of the synaptonemal complex. Using single-molecule imaging and biochemical modeling, Rog and Dernburg demonstrated that the central region of the complex exhibits liquid-like properties, assembling as a thin film that “wets” and spreads along chromosome axes to form a continuous interface between homologs [7]. This liquid–liquid phase separation model explains how the complex can rapidly extend and distribute symmetrically along homologous chromosomes. Further studies, by analyzing the electrostatic surface potential of meiotic chromosome axis proteins (such as HIM-3), have revealed how their molecular properties directly influence the recruitment of synaptonemal complex components and the overall structural stability (Figure 3). Beyond serving as a scaffold to promote crossover formation, the synaptonemal complex may also actively regulate crossover number and distribution. Experimental evidence suggests that it could mediate the transmission of crossover interference signals, whereby one crossover suppresses additional events nearby. Nevertheless, the underlying mechanisms remain unclear and warrant further investigation.
3.1.3 Regulation of crossover number
The AAA+ ATPase PCH-2 serves as a conserved negative regulator of CO formation [8]. It prevents multiple COs from forming on the same chromosome by destabilizing excess CO intermediates, acting both early to limit CO fate adoption and later to eliminate supernumerary intermediates. Loss of PCH-2 leads to increased CO number and loss of interference. While its exact targets in worms are unclear, parallels with other species suggest PCH-2 may remodel HORMA-domain proteins or recombination complexes.
In summary, meiotic progression in C. elegans is driven by a coordinated program: PC-guided homolog recognition, SC-mediated synapsis, and crossover designation by MutSγ/COSA-1, all under surveillance by regulators like PCH-2. These mechanisms ensure one CO per homolog pair and exemplify how conserved and worm-specific strategies together ensure genomic fidelity during meiosis.
3.2. Homolog separation and spindle assembly
3.2.1 Acentrosomal spindle assembly
Oocytes of C. elegans assemble the meiotic spindle without canonical centrosomes. In meiosis I, spindle assembly is chromosome-driven [9]. Following nuclear envelope breakdown, chromatin-associated factors (e.g., Ran-GTP) nucleate microtubules, which motor proteins organize into a bipolar array. Dynein and the kinesin-14 KLP-18, together with proteins such as BMK-1, focus microtubule minus ends to establish two poles despite the absence of centrosomes. Perturbation of these activities (e.g., klp-18 loss) yields mono- or multipolar spindles and blocks homolog segregation.
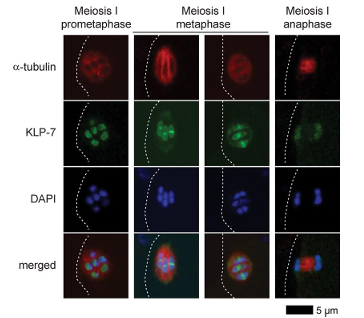
KLP-7 (kinesin-13) limits microtubule abundance and suppresses ectopic pole formation by acting at kinetochores. In klp-7 mutants, spindles accumulate excess microtubules and frequently become multipolar, leading to chromosome mis-pulling during anaphase I. Thus, controlled microtubule turnover and correction of improper attachments are essential for acentrosomal spindle bipolarity. Studies on the dynamic localization of KLP-7 show that it is precisely distributed on the spindle and chromosomes during early to mid-prophase I, metaphase, and anaphase of meiosis I. These dynamic changes are critical for regulating spindle microtubule length and maintaining spindle bipolarity (Figure 4).
3.2.2 Orientation of holocentric chromosomes
The holocentric architecture of C. elegans chromosomes imposes distinctive mechanics. Each bivalent bears a single distal crossover, and kinetochores extend along the chromatid axis. In metaphase I, kinetochores form a cup-like configuration around the short arms [5]; microtubules attach laterally, producing co-orientation of sister chromatids toward one pole and bi-orientation of homologs toward opposite poles. At anaphase I, AIR-2 (Aurora B) enriches on short arms, phosphorylates cohesin (e.g., REC-8), and recruits separase (SEP-1) to release short-arm cohesion, while long-arm cohesion is protected by SGO-1 until meiosis II.
Spindle microtubules often form parallel bundles along chromosome arms, creating channels for lateral transport toward the metaphase plate. KLP-19 at the short-arm ring complex generates plus-end-directed forces that drive congression; its loss abolishes outward movement. The ring complex (AIR-2, BUB-1, KLP-19, etc.) persists into early anaphase I and disassembles after homolog separation, consistent with a regulatory role.
3.2.3 Key factors: KLP-7, MEI-1/2, and AIR-2
Beyond KLP-7, the katanin complex MEI-1/MEI-2 promotes formation of short, focused microtubule bundles required for compact meiotic spindles; in mei-1 or mei-2 mutants, spindles fail to organize and meiosis I arrests. MEI-1 is rapidly eliminated post-meiosis (via CUL-3) to permit normal embryonic mitosis. AIR-2, the Aurora B kinase, is indispensable for short-arm cohesion release and ring-complex assembly in meiosis I and relocalizes between sister chromatids in meiosis II to ensure their separation. Together, these factors integrate conserved and nematode-specific mechanisms to sculpt the spindle and execute accurate chromosome segregation.
3.3. Meiotic checkpoints and regulatory networks
3.3.1 Synapsis (pachytene) checkpoint
In C. elegans, a stringent pachytene checkpoint monitors homolog pairing and recombination. When unsynapsed homologs persist—due to defects such as synaptonemal complex (SC) mutations or absence of Spo11-induced crossovers—apoptosis is triggered to prevent defective gamete production. This checkpoint involves conserved spindle assembly checkpoint (SAC) proteins and meiosis-specific factors, notably MAD-1/MAD-2 and BUB-3 (MDF-1, MDF-2, and BUB-3 in worms) [10]. Mutations in these genes disrupt checkpoint activation, allowing meiotic progression despite synapsis defects.
The AAA+ ATPase PCH-2 is another key regulator. In SC mutants, loss of PCH-2 suppresses arrest only when Spo11-induced double-strand breaks (DSBs) are present, suggesting that PCH-2 primarily monitors recombination intermediates. By contrast, MAD-1/MAD-2 likely respond to structural features of the SC itself. Distinct SC mutants rely on different checkpoint branches: for example, syp-4 mutants require MDF-1 but not PCH-2, whereas syp-3 mutants arrest independently of PCH-2. These observations indicate at least two parallel pathways—one MAD-1/MAD-2–dependent and synapsis-focused, and another PCH-2–dependent and recombination-focused—ensuring high-fidelity meiotic control.
3.3.2 Spindle assembly checkpoint (SAC)
In C. elegans oocytes, a SAC akin to the mitotic SAC operates but is weaker. Although MDF-2 transiently accumulates on unattached kinetochores, severe spindle defects often fail to fully arrest meiosis [11]. This attenuated response likely reflects the biological priority of completing oogenesis. In extreme cases (e.g., monopolar spindles), SAC activity delays anaphase onset and ring complex disassembly. In male spermatocytes, the SAC is even less effective; microtubule inhibitors fail to arrest meiosis, often resulting in aneuploid sperm. These sex-specific differences likely reflect evolutionary trade-offs favoring reproductive output.
3.3.3 CHK-2 and cell cycle integration
CHK-2, a meiosis-specific kinase homologous to ATR/ATM substrates, coordinates early meiotic events. It is activated upon entry into prophase I and promotes pairing center–dependent chromosome movement, SC assembly, and DSB formation. As pachytene progresses, CHK-2 is inactivated by CDK-1–dependent phosphorylation of SC components (e.g., SYP-1 Thr452), which recruits PLK-2 to displace CHK-2 from the chromosomes. This feedback ensures timely exit from pachytene once homologous synapsis is complete.
Oocytes arrest at diakinesis until ovulation, maintained by CDK-1 activity. Upon receiving sperm-derived MSP signals, a MAPK pathway activates CDC-25.3, which in turn activates CDK-1 to resume meiosis [1]. Hence, meiotic progression is tightly regulated by CHK-2, CDK-1, PLK-2, and SAC components, ensuring fidelity and timing.
3.3.4 Network integration and systems-level insights
Meiotic progression in C. elegans is governed by an integrated network linking chromosomal events (synapsis, recombination) to cell cycle regulators. Successful pairing and recombination deactivate CHK-2 and PCH-2, allowing CDK-1 to promote meiotic division. In contrast, defective events activate MAD-1/MAD-2 and SAC components to delay or arrest progression, often inducing apoptosis [8].
The unique transparency and experimental accessibility of C. elegans enable dissection of these pathways within a single organism. Recent advances, including quantitative phosphoproteomics and multi-mutant genetic analyses, are beginning to reconstruct these regulatory networks. Such integrative approaches promise a systems-level understanding of meiotic quality control and its coordination with cell cycle dynamics.
4. Conclusion and outlook
Research in C. elegans has provided critical mechanistic insights into meiosis. Key discoveries include the chromosome-specific pairing center for homolog recognition, holocentric chromosome segregation via ring complexes and lateral microtubule attachments, and checkpoint pathways such as the MAD-1/MAD-2–mediated synapsis checkpoint and PCH-2–regulated crossover control. These studies have collectively built a gene-to-organelle framework for meiotic progression in worms.
It is now understood that homolog pairing relies on specialized chromosomal domains and nuclear movements; crossover number is tightly controlled by dedicated protein modules; and acentrosomal spindles assemble via chromosome-derived cues. Multiple surveillance mechanisms, including CHK-2 and SAC components, ensure genomic integrity throughout gametogenesis. Together, these mechanisms reveal both conserved principles of meiosis and species-specific adaptations.
Despite its strengths, the worm model has limitations. First, C. elegans is self-fertilizing and holocentric, differing from outcrossing, monocentric organisms like mammals. Hence, not all mechanisms are directly translatable. Second, research has predominantly focused on oogenesis, with male meiosis understudied due to limited male germ cells. This leaves gaps in understanding processes such as spermatocyte chromatin remodeling and sex-specific checkpoint control. Third, its small genome and cell size, while advantageous for microscopy and genetics, limit high-throughput biochemical applications. These challenges underscore the need for complementary models, rather than diminishing the worm’s utility.
Looking forward, C. elegans remains a promising system for meiosis research. Emerging multi-omics technologies—such as single-cell RNA-seq and Hi-C—will enable high-resolution mapping of gene expression and 3D genome organization across meiotic stages. Advanced imaging methods, including light-sheet microscopy, combined with machine learning, will facilitate real-time, quantitative analyses of dynamic events like synaptonemal complex (SC) assembly and chromosome movement. Computational pipelines may detect phenotypic subtleties from large datasets with unprecedented sensitivity.
Furthermore, cross-species comparisons and synthetic biology approaches offer translational potential. Comparative analyses with Drosophila and mouse mutants can identify conserved versus lineage-specific pathways. CRISPR-based "humanized" worms—expressing human orthologs in place of worm genes—provide a unique platform for functional interrogation within a live germline context.
Finally, insights from C. elegans meiosis extend beyond basic biology. Mechanistic understanding of germline quality control and apoptosis in worms may inform studies of fertility, genome maintenance, and developmental disorders in higher organisms.
In conclusion, C. elegans has proven indispensable for dissecting the molecular and cellular logic of meiosis. With ongoing technological integration and comparative studies, this model organism will continue to illuminate core principles of heredity and reproduction.
References
[1]. Bohr T, Ashley G, Eggleston E, Firestone K, Bhalla N. Synaptonemal Complex Components Are Required for Meiotic Checkpoint Function in Caenorhabditis elegans. Genetics 2016; 204(3): 987-97.
[2]. Pintard L, Bowerman B. Mitotic Cell Division in Caenorhabditis elegans. Genetics 2019; 211(1): 35-73.
[3]. Woodruff GC, Teterina AA. Degradation of the Repetitive Genomic Landscape in a Close Relative of Caenorhabditis elegans. Mol Biol Evol 2020; 37(9): 2549-67.
[4]. Özdemir I, Steiner FA. Transmission of chromatin states across generations in C. elegans. Seminars in Cell & Developmental Biology 2022; 127: 133-41.
[5]. Horton HH, Divekar NS, Wignall SM. Newfound features of meiotic chromosome organization that promote efficient congression and segregation in Caenorhabditis elegans. MOLECULAR BIOLOGY OF THE CELL 2022; 33(14).
[6]. Bahrami AK, Zhang Y. When females produce sperm: genetics of C. elegans hermaphrodite reproductive choice. G3 (Bethesda) 2013; 3(10): 1851-9.
[7]. Gordon SG, Rodriguez AA, Gu Y, Corbett KD, Lee CF, Rog O. The synaptonemal complex aligns meiotic chromosomes by wetting. Science Advances; 11(9): eadt5675.
[8]. Patel B, Grobler M, Herrera A, Logari E, Ortiz V, Bhalla N. The conserved ATPase PCH-2 controls the number and distribution of crossovers by antagonizing their formation in Caenorhabditis elegans. eLife 2025; 13: RP102409.
[9]. Connolly AA, Sugioka K, Chuang CH, Lowry JB, Bowerman B. KLP-7 acts through the Ndc80 complex to limit pole number in C. elegans oocyte meiotic spindle assembly. J Cell Biol 2015; 210(6): 917-32.
[10]. Bohr T, Nelson CR, Klee E, Bhalla N. Spindle assembly checkpoint proteins regulate and monitor meiotic synapsis in C. elegans. Journal of Cell Biology 2015; 211(2): 233-42.
[11]. Chen SY, Cheng PW, Peng HF, Wu JC. C. elegans spermatocyte divisions show a weak spindle checkpoint response. JOURNAL OF CELL SCIENCE 2024; 137(6).
Cite this article
Li,B. (2025). Caenorhabditis Elegans as a Model System for Meiosis Research: Advantages and Recent Progress. Theoretical and Natural Science,138,25-34.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of ICBioMed 2025 Symposium: Computational Modelling and Simulation for Biology and Medicine
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Bohr T, Ashley G, Eggleston E, Firestone K, Bhalla N. Synaptonemal Complex Components Are Required for Meiotic Checkpoint Function in Caenorhabditis elegans. Genetics 2016; 204(3): 987-97.
[2]. Pintard L, Bowerman B. Mitotic Cell Division in Caenorhabditis elegans. Genetics 2019; 211(1): 35-73.
[3]. Woodruff GC, Teterina AA. Degradation of the Repetitive Genomic Landscape in a Close Relative of Caenorhabditis elegans. Mol Biol Evol 2020; 37(9): 2549-67.
[4]. Özdemir I, Steiner FA. Transmission of chromatin states across generations in C. elegans. Seminars in Cell & Developmental Biology 2022; 127: 133-41.
[5]. Horton HH, Divekar NS, Wignall SM. Newfound features of meiotic chromosome organization that promote efficient congression and segregation in Caenorhabditis elegans. MOLECULAR BIOLOGY OF THE CELL 2022; 33(14).
[6]. Bahrami AK, Zhang Y. When females produce sperm: genetics of C. elegans hermaphrodite reproductive choice. G3 (Bethesda) 2013; 3(10): 1851-9.
[7]. Gordon SG, Rodriguez AA, Gu Y, Corbett KD, Lee CF, Rog O. The synaptonemal complex aligns meiotic chromosomes by wetting. Science Advances; 11(9): eadt5675.
[8]. Patel B, Grobler M, Herrera A, Logari E, Ortiz V, Bhalla N. The conserved ATPase PCH-2 controls the number and distribution of crossovers by antagonizing their formation in Caenorhabditis elegans. eLife 2025; 13: RP102409.
[9]. Connolly AA, Sugioka K, Chuang CH, Lowry JB, Bowerman B. KLP-7 acts through the Ndc80 complex to limit pole number in C. elegans oocyte meiotic spindle assembly. J Cell Biol 2015; 210(6): 917-32.
[10]. Bohr T, Nelson CR, Klee E, Bhalla N. Spindle assembly checkpoint proteins regulate and monitor meiotic synapsis in C. elegans. Journal of Cell Biology 2015; 211(2): 233-42.
[11]. Chen SY, Cheng PW, Peng HF, Wu JC. C. elegans spermatocyte divisions show a weak spindle checkpoint response. JOURNAL OF CELL SCIENCE 2024; 137(6).









