1. Introduction
Mevalonate (MVA), also known as 3,5-dihydroxy-3-methylpentanoic acid, is a six-carbon organic acid that plays a pivotal role in biological metabolism (Figure 1). Under acidic conditions, it can spontaneously cyclize to form mevalonolactone [1]. Widely distributed in the metabolic networks of both prokaryotes and eukaryotes [2], MVA serves as a core intermediate in the MVA pathway, linking primary carbon metabolism to the biosynthesis of diverse bioactive molecules [3]. The discovery and characterization of MVA trace back to landmark studies in the mid-20th century. In 1956, Wolf and colleagues first identified MVA in alcoholic fermentation waste liquor, noting its ability to replace acetate as a growth factor for Lactobacillus heterohiochi, highlighting its potential role in microbial metabolism [4]. Subsequently, Japanese researcher Gakuzo Tamura isolated a compound termed "hiotic acid" from culture of Aspergillus oryzae, which was later identified as MVA [5]. This work further revealed MVA′s involvement in microbial growth regulation and laid the groundwork for its biochemical characterization.
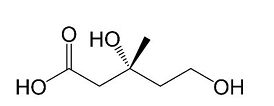
Biologically, MVA is indispensable as a precursor for the synthesis of a vast array of natural products. As the direct precursor of isopentenyl pyrophosphate and dimethylallyl pyrophosphate, the basic building blocks of all terpenoids, it contributes to the biosynthesis of sterols (e.g., cholesterol), steroid hormones, carotenoids, paclitaxel, and biofuel precursors [6-9]. Its role in maintaining cellular homeostasis and supporting the production of high-value compounds underscores its significance in fields ranging from medicine to industrial biotechnology [10].
The production of MVA has historically relied on two main approaches: chemical synthesis and biosynthesis. Chemical synthesis, though feasible, suffers from high costs due to expensive reagents and catalysts, harsh reaction conditions (e.g., high temperature, pressure, or extreme pH), and environmental concerns from toxic by-products [11, 12]. In contrast, biosynthesis driven by microbial metabolic engineering utilizes renewable carbon sources and engineered strains expressing key enzymes of the MVA pathway. This method offers advantages including sustainable substrate availability and environmental compatibility. Commonly used microbial chassis for MVA biosynthesis include Saccharomyces cerevisiae [13], Escherichia coli [14, 15], and Pseudomonas [16, 17], among others.
The MVA pathway was first identified in yeast and animal systems by Bloch′s research group and Lynen et al. in 1958 [18], and it was subsequently found to be associated with ergosterol synthesis in yeast [19]. This pathway is conserved across a wide range of organisms [20], including fungi, mammals, higher plants, and certain bacteria such as Staphylococcus aureus [21], Enterococcus faecalis [22], and Lactobacillus casei [23]. Notably, the MVA pathway of E. faecalis is widely exploited for microbial MVA production due to its efficient enzyme activities [24]. The pathway initiates with acetyl-CoA, which is converted to acetoacetyl-CoA by acetoacetyl-CoA thiolase. This intermediate then condenses with another molecule of acetyl-CoA to form 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA) under the catalysis of HMG-CoA synthase (HMGS). Finally, HMG-CoA is reduced to MVA by HMG-CoA reductase (HMGR) using NADPH as a cofactor (Figure 2). In S. cerevisiae, these key enzymes are encoded by ERG10 (acetoacetyl-CoA thiolase), ERG13 (HMGS), and HMG1/HMG2 (HMGR) [25]; in E. faecalis, acetoacetyl-CoA thiolase and HMGR are encoded by mvaE, while HMGS is encoded by mvaS [26].
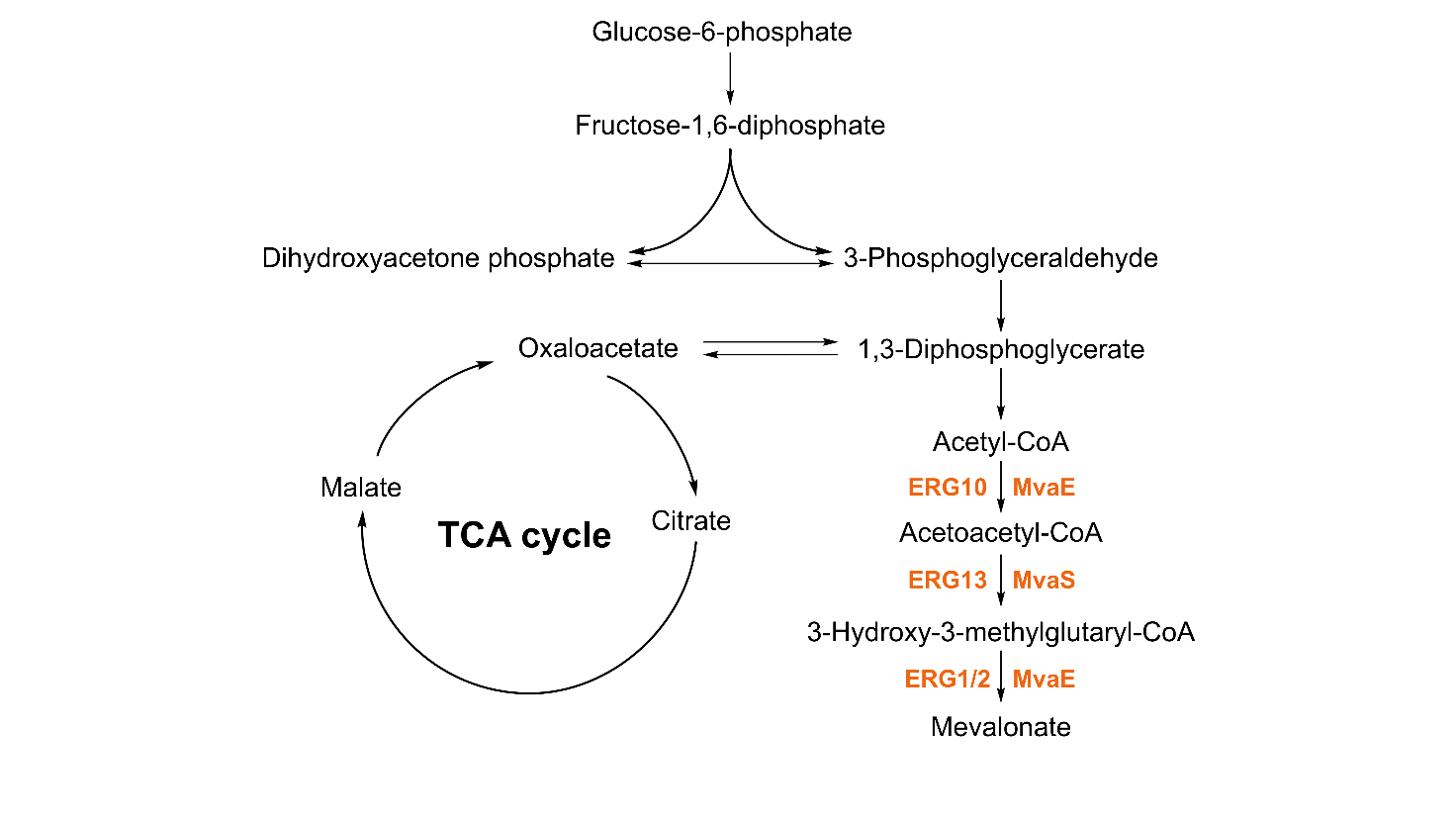
To enhance MVA production, metabolic engineering strategies have focused on optimizing the expression of these key enzymes or heterologously expressing the entire pathway in hosts. Such modifications aim to boost flux through the MVA pathway, representing the core of microbial MVA synthesis and a critical foundation for improving yields. As research into MVA biosynthesis advances, further understanding its metabolic regulation and pathway engineering will be pivotal for unlocking its full potential in industrial applications.
2. Biosynthesis of MVA in microbial hosts
Microbial hosts have emerged as robust cell factories for MVA biosynthesis, with extensive efforts focused on metabolic engineering strategies to enhance pathway flux, precursor supply, and cofactor balance, while minimizing carbon loss to byproducts. This section summarizes the progress in MVA production using major microbial chassis, including yeasts (S. cerevisiae and Yarrowia lipolytica), bacteria (E. coli, Pseudomonas putida), and methylotrophs (Methylobacterium extorquens), highlighting strain engineering strategies, key metabolic modifications, and the resulting improvements in production. Increases in MVA production achieved by these studies are summarized in Table 1.
2.1. Yeast hosts
Yeasts are widely employed as microbial chassis for MVA production, with S. cerevisiae and Y. lipolytica being the most extensively studied due to their favorable biological traits and engineering feasibility [27, 28].
2.1.1. S. cerevisiae
As a Generally Recognized as Safe (GRAS) organism, S. cerevisiae possesses several advantageous traits, including well-established fermentation protocols, high biocompatibility, rapid growth kinetics, ease of product recovery, a well-characterized genetic background, and amenability to genetic manipulation [27, 28]. Wegner et al. achieved significant enhancements in MVA production by stably integrating E. faecalis MVA pathway key enzymes (encoded by mvaE and mvaS) and a feedback-insensitive acetyl-CoA synthetase from Salmonella enterica into S. cerevisiae genomic sites, which synergistically boosted acetyl-CoA supply and MVA synthesis flux. They further minimized metabolic flux diversion by regulating squalene synthase (ERG9) via the pMET3 promoter, while reinforcing acetyl-CoA biosynthesis through pantothenate kinase (CAB1) overexpression combined with pantothenate supplementation. These modifications yielded 3830 120 mg/L MVA in shake-flask cultures, with fed-batch bioreactors achieving 13.3 0.5 g/L [29]. Rodriguez et al. enhanced cytoplasmic acetyl-CoA pools by manipulating citrate metabolism in S. cerevisiae. They knocked out mitochondrial NAD-dependent isocitrate dehydrogenase (IDH1) to shunt citrate from the TCA cycle to the cytoplasm, and overexpressed Aspergillus nidulans ATP citrate lyase for efficient citrate-to-acetyl-CoA conversion. Concomitantly, they introduced E. faecalis MVA pathway genes (mvaE, mvaS) to boost upstream flux and replaced the ERG12 promoter with the copper-regulated PCTR3 to block downstream metabolism, yielding 30 mg/L MVA under low-nitrogen conditions [13]. Jakočiūnas et al. used the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 to engineer S. cerevisiae for enhanced MVA production, targeting five key loci: rox1 (repressor of MVA pathway genes), ypl062w and yjl064w (uncharacterized genes promoting MVA flux), bts1 (branch-point enzyme for isoprenoid synthesis) as well as the erg9 promoter (truncated to reduce ergosterol flux). They generated 31 strains with single to quintuple mutations, identifying 20 with higher MVA titers than the wild type. The top quintuple mutant (disruptions of rox1, ypl062w, yjl064w, bts1 plus truncated erg9 promoter) showed a 41.5-fold increase [25]. Fink et al. enhanced MVA production via a coiled-coil domain-mediated metabolic enzyme clustering strategy: key MVA pathway enzymes (ERG10, HMGS, and tHMGR) were fused with the P3:GCN:P4 coiled-coil domain to form enzyme complexes, thereby boosting directed metabolic flux. After 72 h of fermentation, this strategy increased MVA accumulation by 8.8-fold compared to strains expressing non-scaffolded enzymes [30].
2.1.2. Y. lipolytica
Y. lipolytica, another promising yeast chassis, has also been engineered for efficient MVA production. Zhang et al. optimized MVA biosynthesis in Y. lipolytica via a stepwise engineering approach [3]. Initially, they downregulated ERG12 (to reduce MVA consumption) through promoter replacement, combined with overexpression and copy number amplification of HMGR and ERG13, achieving a shake-flask titer of 4.16 g/L. To enhance acetyl-CoA supply, they overexpressed the citrate transporter YHM2 and ATP-citrate lyase (ACL1/2), while deleting mitochondrial isocitrate dehydrogenases (IDH1/2) to block TCA cycle flux diversion. Introduction of a high-activity citrate lyase from A. nidulans (AnACLa/b) further increased the titer to 5.25 g/L. Finally, deletion of the citrate efflux protein CEX1 eliminated overflow metabolism, resulting in a fed-batch MVA titer of 101 g/L with a yield of 0.255 g/g glucose and this represents the highest MVA production titer reported in microbial systems to date.
2.2. E. coli hosts
E. coli, one of the most extensively studied bacterial species and a major model microorganism, possesses advantages such as a high growth rate, simple culture requirements, and well-defined genetic and physiological characteristics [31]. The conventional metabolic engineering methods for MVA production in E. coli typically involve introducing MVA pathway-related genes from different microorganism sources like E. faecalis and L. casei.
Satowa et al. enhanced the MVA synthesis pathway by introducing the mvaE and mvaS genes from E. faecalis and synergistically overexpressing the endogenous atoB gene (encoding acetyl-CoA acetyltransferase). To further strengthen intracellular acetyl-CoA availability, the gltA gene (encoding citrate synthase) was disrupted to block the TCA cycle, which led to a 7-fold increase in acetyl-CoA levels. This engineered strain produced 8.0 g/L of MVA from 20 g/L glucose [15]. Within this research, an engineered E. coli strain with surface-displayed -glucosidase was also constructed to enable direct utilization of cellobiose. When cultured with 20 g/L cellobiose as the sole carbon source, the gnd (encoding 6-phosphogluconate dehydrogenase)-deficient strain expressing surface-displayed -glucosidase achieved a MVA titer of 5.7 g/L within 24 h. This corresponds to a yield of 0.25 g/g glucose (considering 1 g cellobiose is equivalent to 1.1 g glucose), highlighting that cellobiose serves as a viable alternative carbon source for MVA production in engineered E. coli [15]. Xu et al. co-expressed acs (encoding acetyl CoA synthetase) from E. coli W3110 as well as mvaE and mvaS from E. faecalis in E. coli BL21(DE3) to achieve efficient conversion of acetic acid to acetyl-CoA and subsequent MVA synthesis. A two-stage fermentation strategy was adopted: in the first stage, glucose was used for culture to accumulate biomass; in the second stage, the feed was switched to acetic acid and expression was induced. The MVA titer reached 7.85 g/L within 60 h, with a productivity of 0.13 g/(Lh) in a 5 L fermentor [32]. Dong et al. designed a 0D PP7 RNA scaffold system containing PP7 RNA aptamers, and fused the mvaE and mvaS genes from E. faecalis with RNA-binding domains (RBDs) derived from the PP7 phage coat protein, respectively. Using RNA electrophoretic mobility shift assay, they verified the specific interaction between the PP7 aptamers and RBDs in E. coli BL21(DE3). Evaluation with GFP fluorescent proteins showed that this RNA scaffold system enhanced the co-localization efficiency of enzymes by 375% compared to the control. When applied to MVA synthesis, the engineered strain expressing the 0D PP7 RNA scaffold system achieved an MVA titer of 3.13 g/L after 48 h of induction, which was 84% higher than that of the control strain lacking the scaffold system [26]. Wang et al. constructed a basic synthetic pathway by introducing the mvaES operon from E. faecalis, assembled a phosphoketolase-phosphotransacetylase bypass to enhance acetyl-CoA supply through the non-oxidative glycolysis pathway and reduce carbon loss, and overexpressed gnd to optimize NADPH regeneration. Eventually, in M9Y medium containing 30 g/L glycerol, the final MVA yield in shake-flask fermentation reached 7.21 g/L after 72 h, with a specific yield of 1.36 g/g DCW and a productivity approaching 93.7% of the theoretical value [14]. Kamata et al. focused on optimizing the flux distribution between the Embden-Meyerhof-Parnas (EMP) pathway and the pentose phosphate pathway (PPP) to reduce acetyl-CoA overflow into acetate and enhance MVA production in E. coli. To precisely regulate glycolytic flux, they constructed a strain from E. coli BW25113 with the chromosomal pgi gene (encoding phosphoglucose isomerase, a key enzyme at the branch point of EMP and PPP pathways) knocked out, while expressing pgi via a plasmid under the control of the T5 promoter and Lac operon. This allowed tuning of EMP pathway flux by adjusting isopropyl -D-1-thiogalactopyranoside (IPTG) induction levels, thereby avoiding excessive acetyl-CoA shunting caused by overly high EMP flux. In stationary-phase cultures (under sulfur starvation to minimize biomass synthesis), controlling the EMP pathway flux at 39.7% effectively blocked acetyl-CoA overflow to acetate. This redirection of carbon flux toward MVA synthesis resulted in a carbon yield of 22.1% for MVA, which was 25% higher than that of the strain without flux control (EMP flux of 70.4%). Notably, this optimal flux ratio matched the theoretical value derived from NADPH and acetyl-CoA metabolic balance, confirming that blocking competitive branch pathways (e.g., acetate synthesis) by fine-tuning central carbon flux is an effective strategy to improve MVA yield [33].
Xiong et al. introduced the mvaS and mvaE genes from L. casei into E. coli. In shake-flask fermentation, 14.6 g/L of MVA was produced from 40 g/L glucose. In the scaled-up fermentation in a bioreactor, the strain achieved a productivity of 2 g/(Lh), with a final titer of 88 g/L and a glucose conversion rate of 0.26 g/g [10]. Wang et al. developed a high-efficiency MVA-producing E. coli strain by engineering the MVA pathway through chromosomal integration. They introduced the heterologous atoB, mvaS, and mvaE genes from L. casei into the E. coli chromosome via Red-mediated homologous recombination, with expression controlled by the strong constitutive promoter M1-93. These genes replaced the native adhE and ldhA genes to eliminate by-product formation. To further optimize MVA production, additional metabolic engineering strategies were implemented: deletion of atpFH (encoding H-ATP synthase subunits) to enhance glycolytic flux and accelerate glucose consumption; integration of two copies of the atoB-mvaS-mvaE operon to strengthen downstream flux from acetyl-CoA to MVA; and knockout of sucA (encoding the E1 subunit of 2-oxoglutarate decarboxylase) to block the TCA cycle, redirecting acetyl-CoA toward MVA synthesis. In shake-flask fed-batch fermentation, the engineered strain achieved a MVA titer of 30 g/L from 61 g/L glucose within 48 h, with a yield reaching 86.1% of the maximum theoretical value (0.548 g/g glucose) and a maximum volumetric productivity of 1.01 g/(Lh). The plasmid-free, inducer-independent system exhibits high stability and significant potential for industrial application [34]. To enhance carbon conversion efficiency, Wang et al. developed the EP-bifido pathway in E. coli through the introduction of two key genes: fxpk (encoding a bifunctional phosphoketolase) and fbp (encoding fructose-1,6-bisphosphatase). This design enables the breakdown of fructose 6-phosphate to maximize acetyl-CoA supply from glucose. To further optimize carbon flux, they knocked out edd (encoding 6-phosphogluconate dehydratase) in the Entner-Doudoroff pathway and pfkA (encoding phosphofructose kinase A) in the EMP pathway. This strategy conserved more fructose 6-phosphate for conversion into C2 metabolites, reduced pyruvate flux toward acetyl-CoA, and redirected carbon flow from the EMP pathway to the PPP pathway, thereby increasing NADPH supply. As a result, a high MVA yield of 64.3 mol% was achieved [35]. Building on this foundation, Li et al. further improved MVA production by finely tuning the glycolytic flux ratio of the EP-bifido pathway in E. coli. They optimized the flux distribution between the EMP pathway and PPP by replacing the promoter of zwf (encoding glucose-6-phosphate dehydrogenase) with a series of Anderson promoters of varying strengths (http://parts.igem.org/Promoters/Catalog/Anderson). This modification enhanced PPP flux to boost NADPH supply, leading to a final MVA titer of 11.2 g/L after 72 hours of fermentation, with a glucose-to-MVA molar conversion rate of 62.2% [36]. In the same study, the CRISPR interference (CRISPRi) system was employed to finely downregulate pfkA (encoding phosphofructose kinase A). The MVA yield of the regulated strain was 8.53 g/L, and the conversion rate from glucose reached 68.7% [36].
2.3. P. putida hosts
2.3.1. Crispr-based gene regulation
In the context of MVA biosynthesis in P. putida, the utilization of CRISPR-based gene regulation systems has been validated as a strategy to optimize pathway flux. This typically involves constructing CRISPRa-responsive systems, where key exogenous MVA pathway genes are placed under the control of responsive promoters, combined with induction systems for dynamic transcriptional activation [37]. Additionally, CRISPRi systems are used to target and repress target genes, relieving metabolic inhibition to enhance MVA synthesis efficiency [37, 16]. Kiattisewee et al. placed the mvaES operon from E. faecalis under the control of the CRISPRa-responsive promoter J3-BBa_J23117 to construct the recombinant plasmid, which was introduced into P. putida PPC01, a strain with genomic integration of dCas9/MCP-SoxS. Transcription was activated via J306 scRNA targeting the promoter. Meanwhile, the XylS-Pm induction system was used to regulate dCas9/MCP-SoxS expression, enabling dynamic control. Results showed that the constitutive CRISPRa system produced 402 21 mg/L MVL, while the inducible system yielded 345-397 mg/L under induction with 0.01-1 mM toluic acid [37]. Kim et al. established a CRISPRi-based gene regulation system in P. putida KT2440 [16]. They designed a single-plasmid system where a sgRNA targeting the endogenous repressor gene glpR (encoding a transcriptional repressor of glycerol metabolism) was expressed under a constitutive promoter, while the dCas9 was controlled by the L-rhamnose-inducible PrhaBAD promoter. This CRISPRi plasmid was co-transformed with another plasmid harboring the mvaE and mvaS genes from E. faecalis. Upon induction with 1 mM L-rhamnose, the CRISPRi system specifically repressed glpR, thereby alleviating the transcriptional inhibition of the glpFKRD gene cluster-whose products are responsible for glycerol uptake and catabolism. After 72 hours of cultivation in M9 minimal medium with 4 g/L glycerol as the sole carbon source, the engineered strain achieved an MVA titer of 237 mg/L, representing a 3.3-fold increase compared to the control strain (72 mg/L). Concomitantly, cell density (OD₆₀₀) increased from 1.14 to 2.22, indicating enhanced biomass accumulation alongside improved product synthesis.
2.3.2. Carbon source
Glucose and glycerol, two classical carbon sources, are among the most commonly employed feedstocks for MVA production in P. putida. Zhang et al. demonstrated that fermentations using a recombinant strain derived from P. putida GZT23 with glucose as the carbon source achieved a MVA titer of 470 mg/L after 48 h of shake-flask culture in M9 medium containing 4% glucose [38]. Further scale-up to a 5-L fed-batch fermenter, utilizing a DO-stat strategy (maintaining 20% dissolved oxygen and pH 7.0, with an initial glucose concentration of 20 g/L and automatic feeding upon depletion to maintain supply), resulted in a MVA titer of 5 g/L within 48 h. Yang et al. evaluated 2,3-butanediol (2,3-BDO) as an alternative carbon source, conducting direct comparisons with glucose and glycerol. The BDPP102 strain was cultured in media supplemented with 2,3-BDO, glucose, or glycerol at equimolar carbon concentrations. Their results revealed that 2,3-BDO metabolism proceeds without carbon loss (no CO₂ generation) and is directly converted to acetyl-CoA, yielding 2.21 g/L MVA. In contrast, the metabolic conversion of glucose and glycerol to acetyl-CoA is accompanied by carbon loss, resulting in significantly lower titers of only 0.23 g/L and 0.18 g/L, respectively. These findings highlight 2,3-BDO as a superior carbon source that markedly enhances the efficiency of MVA biosynthesis [17].
2.4. M. extorquens hosts
Zhu et al. engineered M. extorquens AM1 for MVA production from methanol by introducing and optimizing the MVA pathway. They first constructed two heterologous operons: a natural operon (MVE) harboring mvaS and mvaE from E. faecalis, and an artificial operon (MVH) containing hmgcs1 (encoding a high-activity HMGS) from Blattella germanica and tchmgr (encoding a high-activity HMGR) from Trypanosoma cruzi. In shake-flask cultures, these operons yielded 56 mg/L and 66 mg/L of MVA, respectively, with the artificial operon outperforming the natural one. To enhance precursor supply, they introduced phaA (encoding acetyl-CoA acetyltransferase) from Ralstonia eutropha into the MVH operon, constructing the MVT operon. This modification increased MVA titer to 180 mg/L, a 3.2-fold improvement over the natural operon. Further optimization by tuning the ribosome binding site (RBS) strength upstream of phaA to balance pathway flux resulted in a 20% yield increase, with the best strain achieving 215 mg/L in shake flasks. In a 5-L fed-batch fermentation, the optimized strain produced 2.22 g/L of MVA over 310 h, with an overall yield of 28.4 mg MVA/g methanol and a volumetric productivity of 7.16 mg/(Lh) [39]. Liang et al. engineered M. extorquens AM1 for enhanced MVA production by adopting a strategy for sensor-assisted transcriptional regulator engineering. They integrated the RBS-optimized MVA synthesis pathway Mvt-3 at the attTn7 locus and MVA biosensor Sensor-4 at the celABC locus. A key modification was the overexpression of a qscR (encoding a LysR-type transcriptional regulator) mutant (QscR-49) harboring four mutations: Q8* (premature termination at glutamine 8), T61S, N72Y, and E160V. This global mutant regulator redirected carbon flux toward acetyl-CoA, with transcriptional analysis showing upregulation of metabolic genes like fumC (encoding fumarase C) and NADPH generation. In 5-L fed-batch fermentation, the engineered strain produced 2.67 g/L MVA [40].
|
Host strain |
Strategy |
Fermentation scale |
MVA titer (g/L) |
Reference |
|
S. cerevisiae |
||||
|
BY4741 |
1) Deletion of mitochondrial NAD+-dependent isocitrate dehydrogenase (IDH1) to shunt citrate from the TCA cycle to the cytoplasm. 2) Overexpression of A. nidulans ATP citrate lyase for citrate-to-acetyl-CoA conversion. 3) Introduction of E. faecalis MVA pathway genes (mvaE and mvaS). 4) Replacement of the ERG12 promoter with the copper-regulated PCTR3 to block downstream metabolism. |
Shake flask |
0.03 |
[13] |
|
CEN.PK2-1C |
Disruptions of rox1 (repressor of MVA pathway genes), bts1 (branch-point enzyme for isoprenoid synthesis), ypl062w and yjl064w (uncharacterized genes promoting MVA flux) plus truncated erg9 promoter via CRISPR/Cas9. |
Shake Flask |
Not reported |
[25] |
|
CEN.PK2-1C |
1) Integration of MVA pathway genes from E. faecalis and acetyl-CoA synthetase gene from S. enterica into S. cerevisiae genome. 2) Metabolic flux diversion was minimized by regulating squalene synthase (ERG9). 3) Acetyl-CoA biosynthesis reinforcement by pantothenate kinase (CAB1) overexpression and pantothenate supplementation. |
2-L bioreactor |
13.3±0.5 |
[29] |
|
CAR-0002 |
ERG10 (acetyl-CoA acetyl transferase), HMGS and tHMGR (truncated HMGR) were fused with the P3:GCN:P4 coiled-coil domain to form enzyme complexes, thereby boosting directed metabolic flux. |
Shake flask |
Not reported |
[30] |
|
Y. lipolytica |
||||
|
ZG03 |
1) Downregulated ERG12 through promoter replacement. 2) Overexpression and copy number amplification of HMGR and ERG13. 3) Acetyl-CoA supply was enhanced by overexpressing the citrate transporter YHM2 and ATP-citrate lyase ACL1/2 as well as deleting mitochondrial isocitrate dehydrogenases (IDH1/2). 4) Introduction of a citrate lyase from A. nidulans. 5) Deletion of the citrate efflux protein CEX1. |
1.3-L bioreactor |
101 |
[3] |
|
E. coli |
||||
|
BW25113 |
1) Overexpression of atoB. 2) Introduction of mvaE and mvaS from L. casei. |
1.3-L bioreactor |
88 |
[10] |
|
DH5 |
1) Introduction of the mvaES operon from E. faecalis. 2) Enhancement of acetyl-CoA supply via assembling a phosphoketolase-phosphotransacetylase bypass to enhance acetyl-CoA supply. 3) Overexpression of 6-phosphogluconate dehydrogenase to optimize NADPH regeneration. |
Shake flask |
7.21 |
[14] |
|
MG1655 |
1) Introduction of the mvaE and mvaS genes from E. faecalis. 2) Overexpression of the endogenous atoB gene. 3) The gltA gene (encoding citrate synthase) was disrupted to strengthen intracellular acetyl-CoA availability. |
Tube |
5.7 |
[15] |
|
BL21 (DE3) |
The mvaE and mvaS genes from E. faecalis were fused with RBDs via the 0DPP7 RNA scaffold system to achieve spatial co-aggregation of the enzymes. |
Shake flask |
3.13 |
[26] |
|
BL21 (DE3) |
1) Co-expression of acs as well as mvaE and mvaS from E. faecalis. 2) A two-stage fermentation strategy: glucose for biomass accumulation, with the feed switched to acetic acid for expression induction. |
5-L bioreactor |
7.85 |
[32] |
|
BW25113 |
1) Deletion of the chromosomal pgi gene (encoding phosphoglucose isomerase) knocked out. 2) Expression of pgi under the control of the T5 promoter and Lac operon. 3) Tuning EMP pathway flux via adjusted IPTG induction levels to avoid excessive acetyl-CoA shunting from overly high EMP flux. |
Shake flask |
Not reported |
[33] |
|
BW25113 |
1) Chromosomal integrated atoB, mvaS, and mvaE genes from L. casei, driven by promoter M1-93, replacing the native adhE and ldhA genes to eliminate by-product formation. 2) Deletion of atpFH (encoding H⁺-ATP synthase subunits) to enhance glycolytic flux and accelerate glucose consumption. 3) Integration of two copies of the atoB-mvaS-mvaE operon to strengthen downstream flux from acetyl-CoA to MVA. 4) Knockout of sucA (encoding the E1 subunit of 2-oxoglutarate decarboxylase) to block the TCA cycle, redirecting acetyl-CoA toward MVA synthesis. |
Shake flask |
30 |
[34] |
|
BW-P |
Replacement the promoter of zwf (encoding glucose-6-phosphate dehydrogenase) for enhanced PPP flux to boost NADPH supply. |
Shake flask |
11.2 |
[36] |
|
P. putida |
||||
|
KT2440 |
1) Introduction of mvaE and mvaS from E. faecalis. 2) CRISPRi-based inhibition of glpR, thereby alleviating the transcriptional inhibition of glpFKRD-whose products are responsible for glycerol uptake and catabolism. |
Shake flask |
0.237 |
[16] |
|
KT2440 |
1) Introduction of mvaE and mvaS. 2) Overexpression of atoB. 3) 2,3-BDO was utilized as carbon source. |
Shake flask |
2.21 |
[17] |
|
KT2440 |
1) Genomic integration of dCas9/MCP-SoxS. 2) The mvaES operon from E. faecalis was driven by the CRISPRa-responsive promoter J3-BBa_J23117. 3) Transcription of mvaES was activated via J306 scRNA targeting the promoter. |
Shake flask |
0.402 ±0.021 |
[37] |
|
GZT23 |
1) Introduction of atoB, mvaS, and mvaE driven by one J23111 promoter. 2) Regulation of their expression via RBSs of different strengths. 3) Further DO-stat strategy (maintaining 20% dissolved oxygen and pH 7.0, with an initial glucose concentration of 20 g/L and automatic feeding upon depletion to maintain supply). |
5-L bioreactor |
5 |
[38] |
|
M. extorquens |
||||
|
AM1 |
1) Introduction of hmgcs1 (encoding a high-activity HMGS) from B. germanica and tchmgr (encoding a high-activity HMGR) from T. cruzi. 2) Introduction of phaA (encoding acetyl-CoA acetyltransferase) from R. eutropha to enhance precursor supply. 3) Further tuning of the RBS strength upstream of phaA to balance pathway flux. |
5-L bioreactor |
2.22 |
[39] |
|
AM1 |
1) Integration of the RBS-optimized MVA synthesis pathway Mvt-3 at the attTn7 locus and MVA biosensor Sensor-4 at the celABC locus. 2) Overexpression of the mutant QscR-49 regulator to redirect carbon flux toward acetyl-CoA. |
5-L bioreactor |
2.67 |
[40] |
3. Conclusion and prospect
Significant progress has been made in microbial MVA production through metabolic engineering of diverse hosts, including S. cerevisiae, Y. lipolytica, E. coli, P. putida, and M. extorquens. Key strategies such as optimizing MVA pathway enzyme expression, enhancing acetyl-CoA supply, blocking competitive fluxes, and applying synthetic biology tools, such as CRISPR regulation, have yielded remarkable improvements. Notably, Y. lipolytica achieved the highest titer (101 g/L) via citrate metabolism engineering, while E. coli and S. cerevisiae demonstrated industrial potential with titers up to 88 g/L and 13.3 g/L, respectively. Non-model hosts like P. putida and M. extorquens have also shown promise using alternative carbon sources and CRISPR-based regulation. Despite these advances, challenges remain. Future efforts should focus on precision engineering guided by multi-omics to resolve metabolic bottlenecks, developing dynamic regulation systems to balance growth and production, and expanding novel hosts with unique metabolic traits. Additionally, integrating sustainable substrate utilization (e.g., lignocellulose, methanol) and advanced fermentation control will be critical to scaling MVA production economically and environmentally. With synergistic progress in synthetic biology and process optimization, microbial MVA production is poised to become a cornerstone of sustainable biomanufacturing for terpenoids and beyond.
References
[1]. Woollen, B.H., Holme, P.C., Northway, W.J. and Martin, P.D. (2001) Determination of mevalonic acid in human urine as mevalonic acid lactone by gas chromatography-mass spectrometry. Journal of Chromatography B, 760, 179-184.
[2]. Kuzuyama, T. (2002) Mevalonate and nonmevalonate pathways for the biosynthesis of isoprene units. Bioscience Biotechnology and Biochemistry, 66(8), 1619-1627.
[3]. Zhang, G., Ma, Y.R., Huang, M.N., Jia, K.Z., Ma, T., Dai, Z.J. and Wang, Q.H. (2025) Reprograming the carbon metabolism of yeast for hyperproducing mevalonate, a building precursor of the terpenoid backbone. Journal of Agricultural and Food Chemistry, 73, 606-616.
[4]. Wolf, D.E., Hoffman, C.H., Aldrich, P.E., Skeggs, H.R., Wright, L.D. and Folkers, K. (1956) β-Hydroxy-β-methyl δ-valerolactone (divalonic acid), a new biological factor. Journal of the American Chemical Society, 78(17), 4499.
[5]. Tamura, G. (2004) Hiochic acid, a new growth factor for Lactobacillus homohiochi and Lactobacillus heterohiochi (Preliminary report). Journal of General and Applied Microbiology, 50(6), 327-330.
[6]. Faulkner, R. and Jo, Y. (2022) Synthesis, function, and regulation of sterol and nonsterol isoprenoids. Frontiers in Molecular Biosciences, 9, 1006822.
[7]. Zhang, X.K., Wang, D.N., Chen, J., Liu, Z.J., Wei, L.J. and Hua, Q. (2020) Metabolic engineering of β-carotene biosynthesis in Yarrowia lipolytica. Biotechnology Letters, 42(6), 945-956.
[8]. Tong, Y.R., Luo, Y.F. and Gao, W. (2022) Biosynthesis of paclitaxel using synthetic biology. Phytochemistry Reviews, 21(3), 863-877.
[9]. Zheng, Y.N., Liu, Q., Li, L.L., Qin, W., Yang, J.M., Zhang, H.B., Jiang, X.L., Cheng, T., Liu, W., Xu, X. and Xian, M. (2013) Metabolic engineering of Escherichia coli for high-specificity production of isoprenol and prenol as next generation of biofuels. Biotechnology for Biofuels, 6, 57.
[10]. Xiong, M.Y., Schneiderman, D.K., Bates, F.S., Hillmyer, M.A. and Zhang, K.C. (2014) Scalable production of mechanically tunable block polymers from sugar. Proceedings of the National Academy of Sciences of the United States of America, 111(23), 8357-8362.
[11]. Ferraboschi, P., Canevotti, R., Grisenti, P. and Santaniello, E. (1987) A new flexible synthesis of (R, S)-mevalonolactone. Journal of the Chemical Society-Perkin Transactions 1, (11), 2301-2303.
[12]. Lewer, P. and Macmillan, J. (1983) Reinvestigation of a synthesis of (R, S)-mevalonolactone. Journal of the Chemical Society-Perkin Transactions 1, (7), 1417-1420.
[13]. Rodriguez, S., Denby, C.M., Van Vu, T., Baidoo, E.E.K., Wang, G. and Keasling, J.D. (2016) ATP citrate lyase mediated cytosolic acetyl-CoA biosynthesis increases mevalonate production in Saccharomyces cerevisiae. Microbial Cell Factories, 15, 48.
[14]. Wang, Y., Zhou, S.T., Li, R.Y., Liu, Q., Shao, X.X., Zhu, L.Y., Kang, M.K., Wei, G.Y., Kim, S.W. and Wang, C.L. (2022) Reassessing acetyl-CoA supply and NADPH availability for mevalonate biosynthesis from glycerol in Escherichia coli. Biotechnology and Bioengineering, 119(10), 2868-2877.
[15]. Satowa, D., Fujiwara, R., Uchio, S., Nakano, M., Otomo, C., Hirata, Y., Matsumoto, T., Noda, S., Tanaka, T. and Kondo, A. (2020) Metabolic engineering of E. coli for improving mevalonate production to promote NADPH regeneration and enhance acetyl-CoA supply. Biotechnology and Bioengineering, 117, 2153-2164.
[16]. Kim, S.K., Yoon, P.K., Kim, S.J., Woo, S.G., Rha, E., Lee, H., Yeom, S.J., Kim, H., Lee, D.H. and Lee, S.G. (2020) CRISPR interference-mediated gene regulation in Pseudomonas putida KT2440. Microbial Biotechnology, 13, 210-221.
[17]. Yang, J., Im, Y., Kim, T.H., Lee, M.J., Cho, S., Na, J.G., Lee, J. and Oh, B.K. (2020) Engineering Pseudomonas putida KT2440 to convert 2, 3-butanediol to mevalonate. Enzyme and Microbial Technology, 132, 109437.
[18]. Lichtenthaler, H.K. (1999) The 1-deoxy-D-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annual Review of Plant Physiology and Plant Molecular Biology, 50, 47-65.
[19]. Katsuki, H. and Bloch, K. (1967) Studies on the biosynthesis of ergosterol in yeast-Formation of methylated intermediates. Journal of Biological Chemistry, 242(2), 222-227.
[20]. Lombard, J. and Moreira, D. (2011) Origins and early evolution of the mevalonate pathway of isoprenoid biosynthesis in the three domains of life. Molecular Biology and Evolution, 28(1), 87-99.
[21]. Reichert, S., Ebner, P., Bonetti, E.J., Luqman, A., Nega, M., Schrenzel, J., Spröer, C., Bunk, B., Overmann, J., Sass, P., François, P. and Götz, F. (2018) Genetic adaptation of a mevalonate pathway deficient mutant in Staphylococcus aureus. Frontiers in Microbiology, 9, 1539.
[22]. Wilding, E.I., Brown, J.R., Bryant, A.P., Chalker, A.F., Holmes, D.J., Ingraham, K.A., Iordanescu, S., Chi, Y.S., Rosenberg, M. and Gwynn, M.N. (2000) Identification, evolution, and essentiality of the mevalonate pathway for isopentenyl diphosphate biosynthesis in gram-positive cocci. Journal of Bacteriology, 182(15), 4319-4327.
[23]. Alcántara, C. and Zúñiga, M. (2012) Proteomic and transcriptomic analysis of the response to bile stress of Lactobacillus casei BL23. Microbiology-Sgm, 158, 1206-1218.
[24]. Yoon, S.H., Lee, S.H., Das, A., Ryu, H.K., Jang, H.J., Kim, J.Y., Oh, D.K., Keasling, J.D. and Kim, S.W. (2009) Combinatorial expression of bacterial whole mevalonate pathway for the production of β-carotene in E. coli. Journal of Biotechnology, 140, 218-226.
[25]. Jakounas, T., Sonde, I., Herrgård, M., Harrison, S.J., Kristensen, M., Pedersen, L.E., Jensen, M.K. and Keasling, J.D. (2015) Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metabolic Engineering, 28, 213-222.
[26]. Dong, H.G., Liu, C.L., Liu, X.X., Li, Y., Yang, Y.K. and Bai, Z.H. (2022) Increasing mevalonate production mediated by RNA scaffolds system in Escherichia coli. Journal of Biology, 39(4), 18-23.
[27]. Nielsen, J. (2019) Yeast systems biology: model organism and cell factory. Biotechnology Journal, 14, 1800421.
[28]. Ma, C.A.T., Zhang, K.J., Zhang, X.Y., Liu, G.W., Zhu, T.J., Che, Q., Li, D.H. and Zhang, G.J. (2021) Heterologous expression and metabolic engineering tools for improving terpenoids production. Current Opinion in Biotechnology, 69, 281-289.
[29]. Wegner, S.A., Chen, J.M., Ip, S.S., Zhang, Y.F., Dugar, D. and Avalos, J.L. (2021) Engineering acetyl-CoA supply and ERG9 repression to enhance mevalonate production in Saccharomyces cerevisiae. Journal of Industrial Microbiology & Biotechnology, 48, kuab050.
[30]. Fink, T., Stevovic, B., Verwaal, R., Roubos, J.A., Gaber, R., Bencina, M., Jerala, R. and Gradisar, H. (2020) Metabolic enzyme clustering by coiled coils improves the biosynthesis of resveratrol and mevalonate. AMB Express, 10, 97.
[31]. Zhang, X.L., Tervo, C.J. and Reed, J.L. (2016) Metabolic assessment of E. coli as a biofactory for commercial products. Metabolic Engineering, 35, 64-74.
[32]. Xu, X., Xie, M., Zhao, Q., Xian, M. and Liu, H.Z. (2018) Microbial production of mevalonate by recombinant Escherichia coli using acetic acid as a carbon source. Bioengineered, 9(1), 116-123.
[33]. Kamata, K., Toya, Y. and Shimizu, H. (2019) Effect of precise control of flux ratio between the glycolytic pathways on mevalonate production in Escherichia coli. Biotechnology and Bioengineering, 116(5), 1080-1088.
[34]. Wang, J.L., Niyompanich, S., Tai, Y.S., Wang, J.Y., Bai, W.Q., Mahida, P., Gao, T. and Zhang, K.C. (2016) Engineering of a Highly Efficient Escherichia coli Strain for Mevalonate Fermentation through Chromosomal Integration. Applied and Environmental Microbiology, 82(24), 7176-7184.
[35]. Wang, Q., Xu, J.S., Sun, Z.J., Luan, Y.Q., Li, Y., Wang, J.S., Liang, Q.F. and Qi, Q.S. (2019) Engineering an in vivo EP-bifido pathway in Escherichia coli for high-yield acetyl-CoA generation with low CO2emission. Metabolic Engineering, 51, 79-87.
[36]. Li, Y., Xian, H., Xu, Y., Zhu, Y., Sun, Z.J., Wang, Q. and Qi, Q.S. (2021) Fine tuning the glycolytic flux ratio of EP-bifido pathway for mevalonate production by enhancing glucose-6-phosphate dehydrogenase (Zwf) and CRISPRi suppressing 6-phosphofructose kinase (PfkA) in Escherichia coli. Microbial Cell Factories, 20, 32.
[37]. Kiattisewee, C., Dong, C., Fontana, J., Sugianto, W., Peralta-Yahya, P., Carothers, J.M. and Zalatan, J.G. (2021) Portable bacterial CRISPR transcriptional activation enables metabolic engineering in Pseudomonas putida. Metabolic Engineering, 66, 283-295.
[38]. Zhang, L.M., Fan, T.P., Cai, Y.J. and Zheng, X.H. (2024) Production of mevalonate in Pseudomonas putida via tuning the expression of pathway gene. Systems Microbiology and Biomanufacturing, 4, 1162-1173.
[39]. Zhu, W.L., Cui, J.Y., Cui, L.Y., Liang, W.F., Yang, S., Zhang, C. and Xing, X.H. (2016) Bioconversion of methanol to value-added mevalonate by engineered Methylobacterium extorquens AM1 containing an optimized mevalonate pathway. Applied Microbiology and Biotechnology, 100(5), 2171-2182.
[40]. Liang, W.F., Cui, L.Y., Cui, J.Y., Yu, K.W., Yang, S., Wang, T.M., Guan, C.G., Zhang, C. and Xing, X.H. (2017) Biosensor-assisted transcriptional regulator engineering for Methylobacterium extorquens AM1 to improve mevalonate synthesis by increasing the acetyl-CoA supply. Metabolic Engineering, 39, 159-168.
Cite this article
Zhao,S.;Jia,L.;Tang,H. (2025). Strategies for Enhancing the Yield of Mevalonate in Microbial Strains. Theoretical and Natural Science,139,153-164.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of ICBioMed 2025 Symposium: AI for Healthcare: Advanced Medical Data Analytics and Smart Rehabilitation
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Woollen, B.H., Holme, P.C., Northway, W.J. and Martin, P.D. (2001) Determination of mevalonic acid in human urine as mevalonic acid lactone by gas chromatography-mass spectrometry. Journal of Chromatography B, 760, 179-184.
[2]. Kuzuyama, T. (2002) Mevalonate and nonmevalonate pathways for the biosynthesis of isoprene units. Bioscience Biotechnology and Biochemistry, 66(8), 1619-1627.
[3]. Zhang, G., Ma, Y.R., Huang, M.N., Jia, K.Z., Ma, T., Dai, Z.J. and Wang, Q.H. (2025) Reprograming the carbon metabolism of yeast for hyperproducing mevalonate, a building precursor of the terpenoid backbone. Journal of Agricultural and Food Chemistry, 73, 606-616.
[4]. Wolf, D.E., Hoffman, C.H., Aldrich, P.E., Skeggs, H.R., Wright, L.D. and Folkers, K. (1956) β-Hydroxy-β-methyl δ-valerolactone (divalonic acid), a new biological factor. Journal of the American Chemical Society, 78(17), 4499.
[5]. Tamura, G. (2004) Hiochic acid, a new growth factor for Lactobacillus homohiochi and Lactobacillus heterohiochi (Preliminary report). Journal of General and Applied Microbiology, 50(6), 327-330.
[6]. Faulkner, R. and Jo, Y. (2022) Synthesis, function, and regulation of sterol and nonsterol isoprenoids. Frontiers in Molecular Biosciences, 9, 1006822.
[7]. Zhang, X.K., Wang, D.N., Chen, J., Liu, Z.J., Wei, L.J. and Hua, Q. (2020) Metabolic engineering of β-carotene biosynthesis in Yarrowia lipolytica. Biotechnology Letters, 42(6), 945-956.
[8]. Tong, Y.R., Luo, Y.F. and Gao, W. (2022) Biosynthesis of paclitaxel using synthetic biology. Phytochemistry Reviews, 21(3), 863-877.
[9]. Zheng, Y.N., Liu, Q., Li, L.L., Qin, W., Yang, J.M., Zhang, H.B., Jiang, X.L., Cheng, T., Liu, W., Xu, X. and Xian, M. (2013) Metabolic engineering of Escherichia coli for high-specificity production of isoprenol and prenol as next generation of biofuels. Biotechnology for Biofuels, 6, 57.
[10]. Xiong, M.Y., Schneiderman, D.K., Bates, F.S., Hillmyer, M.A. and Zhang, K.C. (2014) Scalable production of mechanically tunable block polymers from sugar. Proceedings of the National Academy of Sciences of the United States of America, 111(23), 8357-8362.
[11]. Ferraboschi, P., Canevotti, R., Grisenti, P. and Santaniello, E. (1987) A new flexible synthesis of (R, S)-mevalonolactone. Journal of the Chemical Society-Perkin Transactions 1, (11), 2301-2303.
[12]. Lewer, P. and Macmillan, J. (1983) Reinvestigation of a synthesis of (R, S)-mevalonolactone. Journal of the Chemical Society-Perkin Transactions 1, (7), 1417-1420.
[13]. Rodriguez, S., Denby, C.M., Van Vu, T., Baidoo, E.E.K., Wang, G. and Keasling, J.D. (2016) ATP citrate lyase mediated cytosolic acetyl-CoA biosynthesis increases mevalonate production in Saccharomyces cerevisiae. Microbial Cell Factories, 15, 48.
[14]. Wang, Y., Zhou, S.T., Li, R.Y., Liu, Q., Shao, X.X., Zhu, L.Y., Kang, M.K., Wei, G.Y., Kim, S.W. and Wang, C.L. (2022) Reassessing acetyl-CoA supply and NADPH availability for mevalonate biosynthesis from glycerol in Escherichia coli. Biotechnology and Bioengineering, 119(10), 2868-2877.
[15]. Satowa, D., Fujiwara, R., Uchio, S., Nakano, M., Otomo, C., Hirata, Y., Matsumoto, T., Noda, S., Tanaka, T. and Kondo, A. (2020) Metabolic engineering of E. coli for improving mevalonate production to promote NADPH regeneration and enhance acetyl-CoA supply. Biotechnology and Bioengineering, 117, 2153-2164.
[16]. Kim, S.K., Yoon, P.K., Kim, S.J., Woo, S.G., Rha, E., Lee, H., Yeom, S.J., Kim, H., Lee, D.H. and Lee, S.G. (2020) CRISPR interference-mediated gene regulation in Pseudomonas putida KT2440. Microbial Biotechnology, 13, 210-221.
[17]. Yang, J., Im, Y., Kim, T.H., Lee, M.J., Cho, S., Na, J.G., Lee, J. and Oh, B.K. (2020) Engineering Pseudomonas putida KT2440 to convert 2, 3-butanediol to mevalonate. Enzyme and Microbial Technology, 132, 109437.
[18]. Lichtenthaler, H.K. (1999) The 1-deoxy-D-xylulose-5-phosphate pathway of isoprenoid biosynthesis in plants. Annual Review of Plant Physiology and Plant Molecular Biology, 50, 47-65.
[19]. Katsuki, H. and Bloch, K. (1967) Studies on the biosynthesis of ergosterol in yeast-Formation of methylated intermediates. Journal of Biological Chemistry, 242(2), 222-227.
[20]. Lombard, J. and Moreira, D. (2011) Origins and early evolution of the mevalonate pathway of isoprenoid biosynthesis in the three domains of life. Molecular Biology and Evolution, 28(1), 87-99.
[21]. Reichert, S., Ebner, P., Bonetti, E.J., Luqman, A., Nega, M., Schrenzel, J., Spröer, C., Bunk, B., Overmann, J., Sass, P., François, P. and Götz, F. (2018) Genetic adaptation of a mevalonate pathway deficient mutant in Staphylococcus aureus. Frontiers in Microbiology, 9, 1539.
[22]. Wilding, E.I., Brown, J.R., Bryant, A.P., Chalker, A.F., Holmes, D.J., Ingraham, K.A., Iordanescu, S., Chi, Y.S., Rosenberg, M. and Gwynn, M.N. (2000) Identification, evolution, and essentiality of the mevalonate pathway for isopentenyl diphosphate biosynthesis in gram-positive cocci. Journal of Bacteriology, 182(15), 4319-4327.
[23]. Alcántara, C. and Zúñiga, M. (2012) Proteomic and transcriptomic analysis of the response to bile stress of Lactobacillus casei BL23. Microbiology-Sgm, 158, 1206-1218.
[24]. Yoon, S.H., Lee, S.H., Das, A., Ryu, H.K., Jang, H.J., Kim, J.Y., Oh, D.K., Keasling, J.D. and Kim, S.W. (2009) Combinatorial expression of bacterial whole mevalonate pathway for the production of β-carotene in E. coli. Journal of Biotechnology, 140, 218-226.
[25]. Jakounas, T., Sonde, I., Herrgård, M., Harrison, S.J., Kristensen, M., Pedersen, L.E., Jensen, M.K. and Keasling, J.D. (2015) Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metabolic Engineering, 28, 213-222.
[26]. Dong, H.G., Liu, C.L., Liu, X.X., Li, Y., Yang, Y.K. and Bai, Z.H. (2022) Increasing mevalonate production mediated by RNA scaffolds system in Escherichia coli. Journal of Biology, 39(4), 18-23.
[27]. Nielsen, J. (2019) Yeast systems biology: model organism and cell factory. Biotechnology Journal, 14, 1800421.
[28]. Ma, C.A.T., Zhang, K.J., Zhang, X.Y., Liu, G.W., Zhu, T.J., Che, Q., Li, D.H. and Zhang, G.J. (2021) Heterologous expression and metabolic engineering tools for improving terpenoids production. Current Opinion in Biotechnology, 69, 281-289.
[29]. Wegner, S.A., Chen, J.M., Ip, S.S., Zhang, Y.F., Dugar, D. and Avalos, J.L. (2021) Engineering acetyl-CoA supply and ERG9 repression to enhance mevalonate production in Saccharomyces cerevisiae. Journal of Industrial Microbiology & Biotechnology, 48, kuab050.
[30]. Fink, T., Stevovic, B., Verwaal, R., Roubos, J.A., Gaber, R., Bencina, M., Jerala, R. and Gradisar, H. (2020) Metabolic enzyme clustering by coiled coils improves the biosynthesis of resveratrol and mevalonate. AMB Express, 10, 97.
[31]. Zhang, X.L., Tervo, C.J. and Reed, J.L. (2016) Metabolic assessment of E. coli as a biofactory for commercial products. Metabolic Engineering, 35, 64-74.
[32]. Xu, X., Xie, M., Zhao, Q., Xian, M. and Liu, H.Z. (2018) Microbial production of mevalonate by recombinant Escherichia coli using acetic acid as a carbon source. Bioengineered, 9(1), 116-123.
[33]. Kamata, K., Toya, Y. and Shimizu, H. (2019) Effect of precise control of flux ratio between the glycolytic pathways on mevalonate production in Escherichia coli. Biotechnology and Bioengineering, 116(5), 1080-1088.
[34]. Wang, J.L., Niyompanich, S., Tai, Y.S., Wang, J.Y., Bai, W.Q., Mahida, P., Gao, T. and Zhang, K.C. (2016) Engineering of a Highly Efficient Escherichia coli Strain for Mevalonate Fermentation through Chromosomal Integration. Applied and Environmental Microbiology, 82(24), 7176-7184.
[35]. Wang, Q., Xu, J.S., Sun, Z.J., Luan, Y.Q., Li, Y., Wang, J.S., Liang, Q.F. and Qi, Q.S. (2019) Engineering an in vivo EP-bifido pathway in Escherichia coli for high-yield acetyl-CoA generation with low CO2emission. Metabolic Engineering, 51, 79-87.
[36]. Li, Y., Xian, H., Xu, Y., Zhu, Y., Sun, Z.J., Wang, Q. and Qi, Q.S. (2021) Fine tuning the glycolytic flux ratio of EP-bifido pathway for mevalonate production by enhancing glucose-6-phosphate dehydrogenase (Zwf) and CRISPRi suppressing 6-phosphofructose kinase (PfkA) in Escherichia coli. Microbial Cell Factories, 20, 32.
[37]. Kiattisewee, C., Dong, C., Fontana, J., Sugianto, W., Peralta-Yahya, P., Carothers, J.M. and Zalatan, J.G. (2021) Portable bacterial CRISPR transcriptional activation enables metabolic engineering in Pseudomonas putida. Metabolic Engineering, 66, 283-295.
[38]. Zhang, L.M., Fan, T.P., Cai, Y.J. and Zheng, X.H. (2024) Production of mevalonate in Pseudomonas putida via tuning the expression of pathway gene. Systems Microbiology and Biomanufacturing, 4, 1162-1173.
[39]. Zhu, W.L., Cui, J.Y., Cui, L.Y., Liang, W.F., Yang, S., Zhang, C. and Xing, X.H. (2016) Bioconversion of methanol to value-added mevalonate by engineered Methylobacterium extorquens AM1 containing an optimized mevalonate pathway. Applied Microbiology and Biotechnology, 100(5), 2171-2182.
[40]. Liang, W.F., Cui, L.Y., Cui, J.Y., Yu, K.W., Yang, S., Wang, T.M., Guan, C.G., Zhang, C. and Xing, X.H. (2017) Biosensor-assisted transcriptional regulator engineering for Methylobacterium extorquens AM1 to improve mevalonate synthesis by increasing the acetyl-CoA supply. Metabolic Engineering, 39, 159-168.









