1. Introduction
Breast cancer, the most prevalent malignant tumour among women, poses a significant threat to female health. Early diagnosis is crucial for improving patient survival rates [1]. Ultrasound examination, with its advantages of being non-invasive, low-cost, and radiation-free, has become a vital tool for breast cancer screening and diagnosis. However, traditional diagnosis relies heavily on physician experience, leading to subjective variations [2], and faces issues such as inadequate detection of minute lesions and low diagnostic consistency [1,3]. In recent years, the application of conventional machine learning techniques in medical image analysis has provided an effective solution to these challenges. By combining artificially designed radiomic features with classification algorithms, it has demonstrated stable auxiliary diagnostic value in the quantitative analysis and disease classification of ultrasound images.
This study focuses on extracting radiomic features within a traditional machine learning framework. By incorporating first-order statistics, morphological, and textural features from breast ultrasound images, along with an original concentric grey-level fitting curve slope feature, we constructed a binary classification diagnostic model using classical classifiers such as SVM, KNN, and random forest. The goal is to enhance the accuracy and objectivity of breast cancer diagnosis, providing clinicians with an interpretable and easily deployable auxiliary diagnostic tool.
2. Review of domestic and foreign research
2.1. International research landscape
Overseas research on traditional machine learning-assisted breast cancer ultrasound diagnosis focuses on feature engineering optimisation and classifier performance enhancement, with an emphasis on clinical utility and multidimensional data fusion.
2.1.1. Radiomics feature extraction and application
Traditional machine learning relies on manually designed radiomic features, with texture and morphological characteristics forming the core of research. Pons et al. [4] proposed a breast lesion segmentation method based on a maximum a posteriori probability framework. By combining the grey-level co-occurrence matrix (GLCM) to extract texture features such as contrast and entropy, and using an SVM classifier for benign-malignant differentiation, they established a reliable feature extraction paradigm for subsequent machine learning models. Huang et al. [5] quantified morphological differences in lesions using features such as edge sharpness and depth-to-width ratio. They found that malignant lesions exhibited significantly higher depth-to-width ratios than benign ones. Combined with a KNN classifier, this approach achieved a diagnostic accuracy of 79.3%, confirming the value of morphological features in binary classification.
2.1.2. Classifier optimisation and clinical integration
FERREA et al. [6] integrated ultrasound texture features (e.g., spatial grey-scale correlation) with clinical indicators (e.g., tumour size, hormone receptor status), using logistic regression to construct a triple-negative breast cancer discrimination model. With an AUC of 0.824, this approach provides quantitative evidence for treatment selection. LEE et al. [7] used a random forest algorithm to predict axillary lymph node metastasis based on morphological and textural features from ultrasound images, achieving an AUC of 0.805. This approach reduced unnecessary invasive examinations by 42%, demonstrating the advantages of traditional machine learning in clinical translation.
2.1.3. Technical challenges and directions for improvement
Overseas research faces challenges of feature redundancy and data heterogeneity. MOON et al. [8] used feature selection algorithms (e.g., recursive feature elimination) to screen key texture features, improving the SVM model's diagnostic accuracy from 68.2% to 75.6%. However, instrument variability across multicentre data reduced model generalisation by 12.3% [9]. Furthermore, subjective differences in manual annotation (approximately 8.7%) also impact feature stability [10] .
2.2. Current state of domestic research
Domestic research, guided by clinical needs, focuses on practical enhancements to traditional machine learning models and the integration of imaging-clinical features, yielding significant outcomes in binary classification diagnostics.Domestic research, guided by clinical needs, focuses on practical enhancements to traditional machine learning models and the integration of imaging-clinical features, yielding significant outcomes in binary classification diagnostics.
2.2.1. Machine learning model performance enhancement
Zhou Yang et al. [1] compared the efficacy of conventional ultrasound with the combined diagnosis of "imaging-omics features + SVM". They found that the combined model demonstrated significantly higher sensitivity (93.33%) and specificity (100%) than conventional diagnosis, with a Kappa agreement of 0.862 against pathological results, confirming the value of traditional machine learning in enhancing diagnostic consistency. Gao Siqi et al. [11] analysed morphological (circularity, perimeter) and textural features (GLCM energy values) distinguishing benign from malignant lesions. Using a random forest model for binary classification, they achieved 78% specificity, providing primary care hospitals with a low-cost diagnostic adjunct.
2.2.2. Integration of radiomics with clinical indicators
Wu Xiao-na et al. [12] constructed a risk prediction model integrating "clinical indicators (palpable texture, age) + ultrasound features (angularity/spicules)", using the LightGBM algorithm for breast cancer risk stratification. With an AUC of 0.847 and validation set sensitivity of 86.11%, this provides an efficient screening tool for high-risk populations. Zhou Jianhua's team at Sun Yat-sen University [13] integrated ultrasound radiomic features with needle biopsy pathology indicators. Using an XGBoost model to distinguish early-stage breast cancer subtypes, they achieved an AUC of 0.900, demonstrating the advantages of multi-dimensional feature fusion.
2.2.3. Clinical translation and application scenarios
Chen Rui et al. [14] developed a KNN-based AI-assisted system for precise classification of BI-RADS 4 nodules by integrating morphological (edge regularity) and textural (grey-level variance) features. Diagnostic accuracy for ≤2cm nodules reached 89.2%, significantly surpassing manual diagnosis (67.5%). Research by Shen Jie et al. [15] showed that "random forest + radiomic features" achieved a 93.3% detection rate for high-risk lesions (BI-RADS category 4 and above), with the system now deployed across 12 primary care hospitals.
2.3. Conclusions
Domestic and international studies confirm that traditional machine learning, combining artificially designed radiomic features with classifiers, can effectively assist breast cancer ultrasound diagnosis. However, challenges persist, including feature redundancy and insufficient multi-centre data compatibility [9]. In comparison, domestic research prioritises adaptability to clinical scenarios, while international studies demonstrate more systematic approaches in feature engineering theory and classifier optimisation, providing methodological references for this study.
3. Experimental
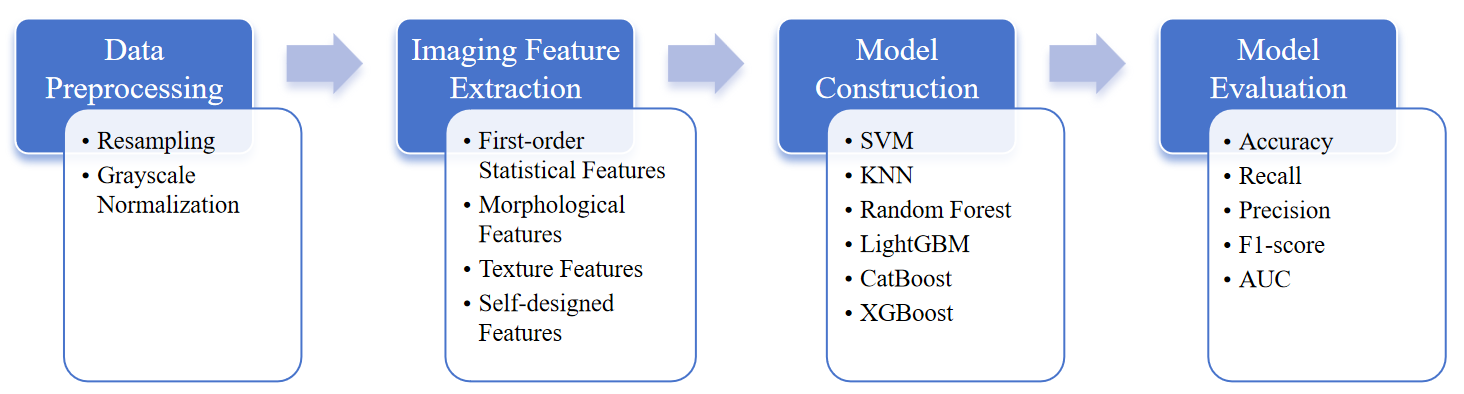
3.1. Research subjects
The experimental data for this study was sourced from the Breast Ultrasound Images Dataset on the Kaggle platform. This open-source dataset comprises 780 breast ultrasound images collected in 2018 from 600 female patients aged 25 to 75. The images are in PNG format with an average size of 500×500 pixels and include ground truth labels categorised into three classes: normal, benign, and malignant. For binary classification, this study categorises the images into two groups: "non-diseased" (normal) and "diseased" (benign + malignant) ( Figure 2,3).
3.2. Radiomics analysis
3.2.1. Data preprocessing
Prior to feature extraction, preprocessing steps including resampling and grey-scale value normalisation were applied to the original images to ensure comparability across different ultrasound devices.Prior to feature extraction, preprocessing steps including resampling and grey-scale value normalisation were applied to the original images to ensure comparability across different ultrasound devices.
3.2.2. Feature extraction
The extracted imaging features encompass four categories:
①First-order statistical features (mean grey value, grey value variance, reflecting overall brightness and internal echo uniformity, respectively);
②Morphometric features (perimeter calculated after threshold segmentation of lesion regions, i.e., contour length of non-zero pixels);
③Texture features (contrast of grey value co-variance matrix, reflecting grey value differences between pixels and lesion heterogeneity);
④ Proprietary feature—slope of concentric grey-level fitting curves.
Extraction process: Determine the lesion region and centre via threshold segmentation; calculate the maximum effective radius and divide it into four concentric circles; compute the mean grey-level for each ring; fit a linear regression model with the ring index as the independent variable and the ring mean grey-level as the dependent variable; the resulting slope constitutes this feature.
Relevant formula:
Maximum effective radius (max_radius), where img_h and img_w denote the height and width of the image, respectively
Mean grey value per ring (ring_mean), where M denotes the total number of pixels within the ring and xₚ represents the grey value of the p th pixel within the ring.
Linear regression model, where k denotes the slope and b denotes the intercept.
3.2.3. Model construction
Multiple classifiers including SVM, KNN, Random Forest, LightGBM, CatBoost, and XGBoost were used for model construction with corresponding parameter tuning. To eliminate feature scale effects, the training set was standardised post-fitting using StandardScaler, followed by standardisation of both training and test sets. Ten-fold stratified cross-validation was applied, with Pipelines ensuring that standardisation occurred only on the training set within each fold to prevent data leakage.
3.2.4. Model evaluation
Model evaluation employed metrics including accuracy, recall, precision, F1 score, and AUC. A three-way table compared metric performance across models. ROC curves (annotated with AUC) and confusion matrices assessed model capability. Feature importance scores were extracted for tree models and visualised via bar charts to illustrate feature significance in predictions.
4. Results
4.1. Overall model performance
Ten-fold stratified cross-validation results indicate varying capabilities among the six models in distinguishing between "non-diseased" and "diseased" images(Table 1). Random Forest and CatBoost demonstrated superior performance: Random Forest achieved an accuracy of 0.689 and AUC of 0.688; CatBoost exhibited the highest recall (0.691) and an F1 score of 0.685. KNN performed relatively weakly, with an accuracy of 0.628 and an AUC of 0.626.
|
Model |
Accuracy |
Recall |
Precision |
F1 |
AUC |
|
SVM |
0.655 |
0.667 |
0.667 |
0.659 |
0.654 |
|
KNN |
0.628 |
0.555 |
0.660 |
0.593 |
0.626 |
|
Random Forest |
0.689 |
0.676 |
0.702 |
0.682 |
0.688 |
|
LightGBM |
0.647 |
0.630 |
0.661 |
0.636 |
0.645 |
|
CatBoost |
0.684 |
0.691 |
0.698 |
0.685 |
0.684 |
|
XGBoost |
0.647 |
0.652 |
0.649 |
0.631 |
0.645 |
4.2. Improvement in model performance through self-created features
Comparing model performance with and without the introduced self-created features (Table 2), all models showed improved performance after incorporating the "slope of concentric grey-scale fitting curves" feature. For example, the LightGBM model’s AUC increased from 0.677 to 0.715 after introducing this feature, with accuracy increasing by 3.0% and recall by 3.9%. For Random Forest, the F1 score increased from 0.671 to 0.682, while AUC remained at 0.688 and recall improved by 2.8%.
|
Model |
Accuracy |
Recall |
Precision |
F1 |
AUC |
|
SVM |
0.670 |
0.714 |
0.674 |
0.685 |
0.670 |
|
KNN |
0.632 |
0.618 |
0.644 |
0.626 |
0.632 |
|
Random Forest |
0.668 |
0.648 |
0.714 |
0.671 |
0.689 |
|
LightGBM |
0.677 |
0.676 |
0.685 |
0.672 |
0.677 |
|
CatBoost |
0.651 |
0.669 |
0.696 |
0.674 |
0.680 |
|
XGBoost |
0.635 |
0.615 |
0.655 |
0.625 |
0.635 |
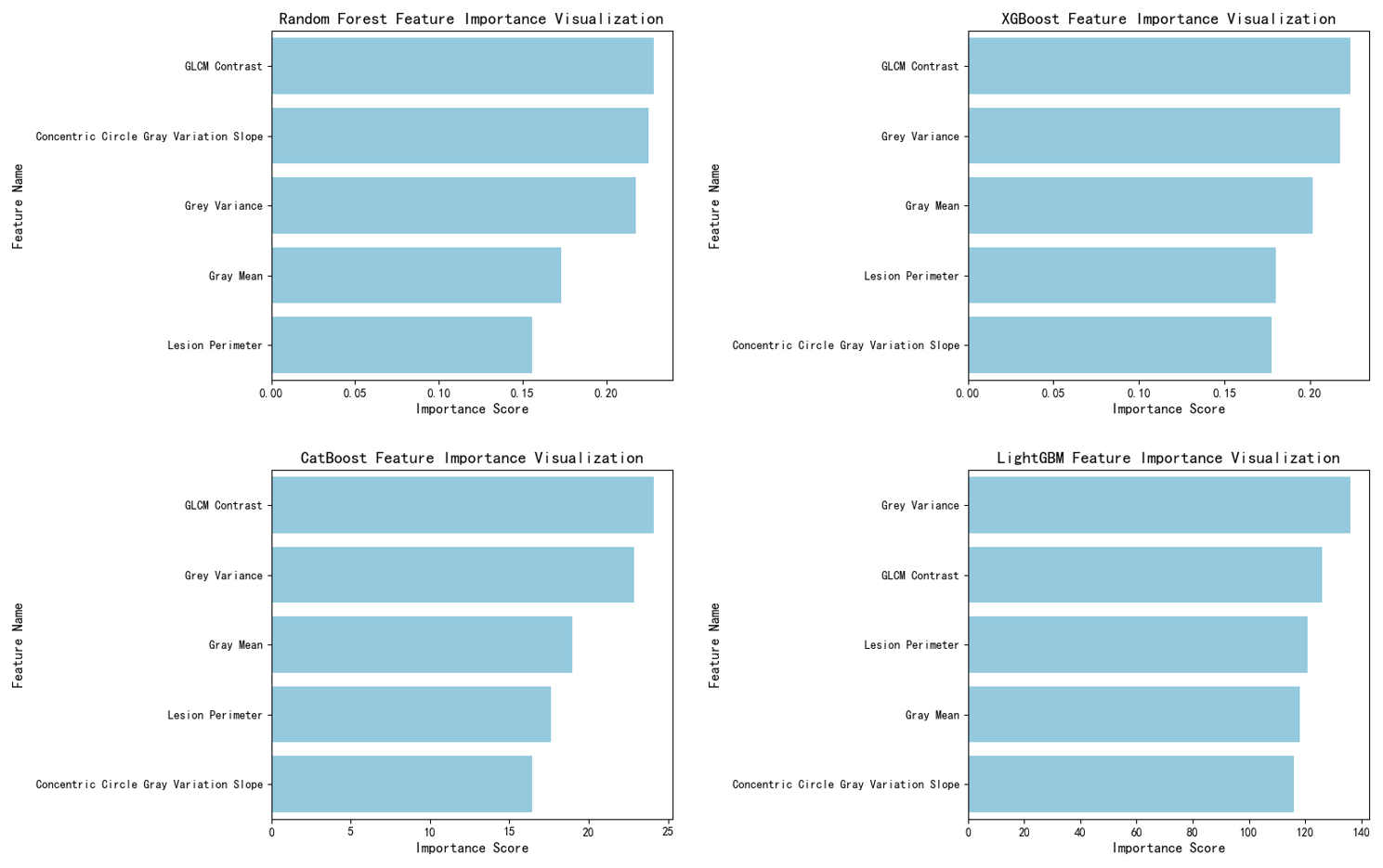
In feature importance analysis, for tree models (Random Forest, LightGBM, CatBoost, XGBoost), the feature importance score for the slope of the concentric grey-scale fitting curve consistently ranked highly ( Figure 4 ). In the Random Forest model, this feature ranked second among all features, indicating its pivotal role in the model's decision-making process by providing effective discriminative information.
4.3. Model performance visualisation results
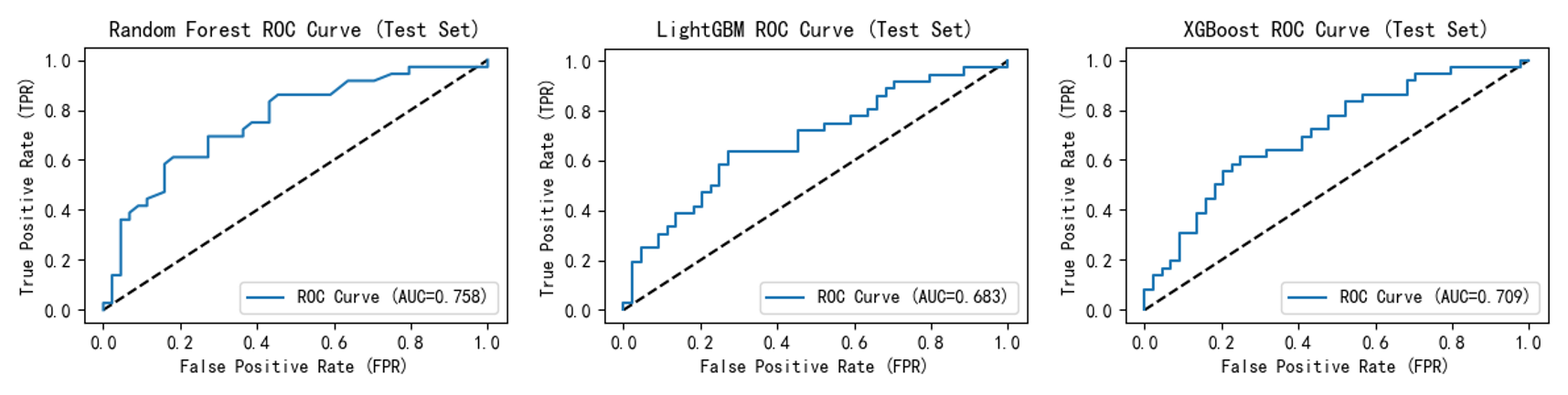
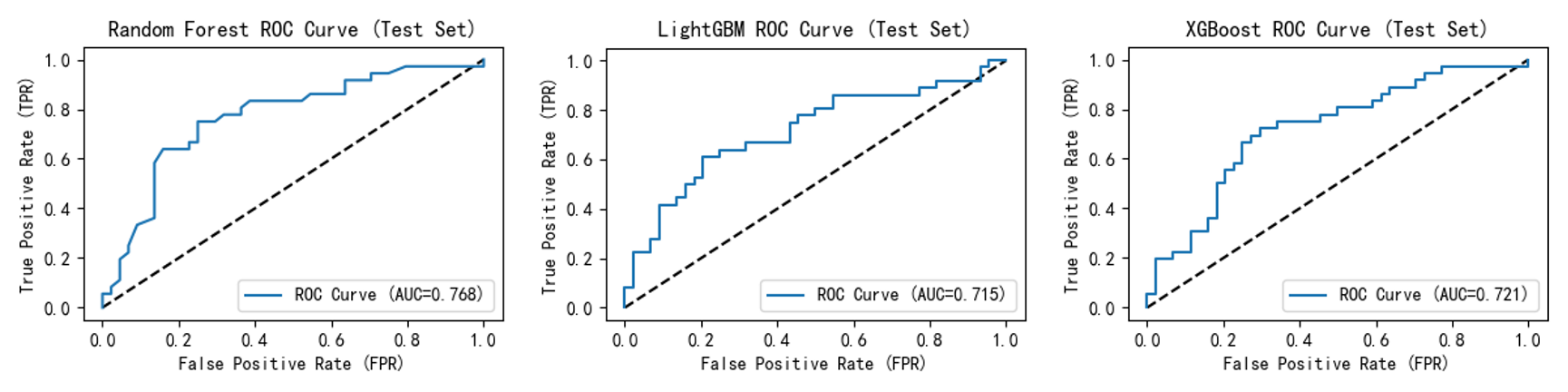
ROC curves ( Figure 5,6 ) show that after incorporating the self-created feature, all model curves shifted closer to the upper-left quadrant. This indicates enhanced model discrimination between "diseased" and "non-diseased" samples, maintaining higher true positive rates at lower false positive rates and improving overall accuracy. For instance, Random Forest improved from 0.758 to 0.768; LightGBM rose from 0.683 to 0.715; and XGBoost's ROC curve AUC increased from 0.709 to 0.721.
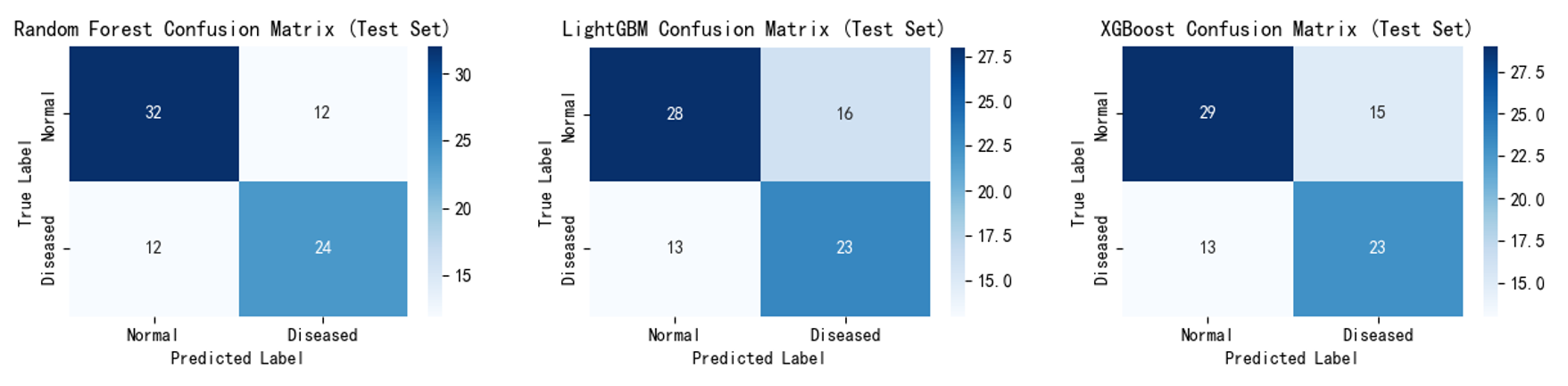
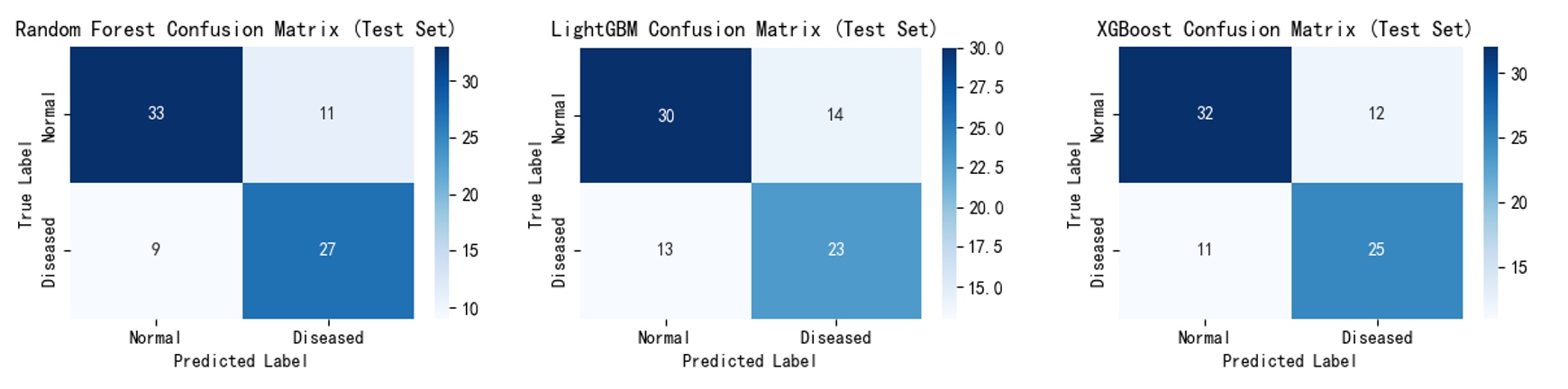
Confusion matrices (Figure 7,8) show that after incorporating the self-created feature, the number of misclassified samples decreased, validating that these features effectively reduce misjudgements and optimise classification performance. For example, Random Forest correctly identified 27 'diseased' samples compared to 24 previously; LightGBM correctly classified 30 'non-diseased' samples compared to 28 previously. XGBoost correctly classified 25 'diseased' samples (up from 23) and 32 'non-diseased' samples (up from 29).
5. Discussion
5.1. Model performance analysis
In this study, Random Forest and CatBoost demonstrated optimal performance, consistent with their advantages in handling non-linear features and high-dimensional data [16,17]. Random Forest mitigates overfitting risk through ensemble decision trees; its high accuracy and AUC indicate that radiomic features (particularly self-created features) contain rich discriminative information. CatBoost's high recall rate holds potential for clinical screening by reducing missed diagnoses. KNN performed weakly, potentially due to noise interference in ultrasound image features and its sensitivity to local sample distributions [18]. SVM, LightGBM, and XGBoost exhibited intermediate performance, with improvements observed after incorporating the novel features, validating the effectiveness of feature design.
5.2. Value and biological significance of the self-created feature
The self-created "concentric circle grey-scale fitting curve slope" feature significantly enhances model performance, primarily by capturing the grey-scale gradient trend from the lesion centre to the periphery. From a biological perspective, uneven proliferation of malignant tumour cells increases internal echo heterogeneity, whereas benign lesions exhibit relatively regular structures with gentler grey-scale gradients [19]. This feature quantifies such gradient differences, supplementing the spatial distribution limitations of traditional texture features, which is consistent with feature importance rankings.
5.3. Limitations and future directions
This study has two limitations: First, data sourced from a single open-access dataset may introduce selection bias, requiring validation with multi-centre clinical data [9]; Second, clinical indicators such as age and medical history were excluded, whereas Wu et al. [12] demonstrated that clinical-imaging fusion further enhances efficacy.
Future research may advance in three directions: ①Expanding the sample size and incorporating multicentre data to enhance model generalisation[9]; ②Integrating clinical indicators with radiomic features to explore deep association mechanisms [13]; ③Optimising proprietary feature extraction algorithms, such as refining concentric circle segmentation rules through morphological operations to improve sensitivity for minute lesions.
6. Conclusion
This study confirms the application potential of radiomics features in breast ultrasound diagnosis of breast cancer. Specifically, the self-created "slope of the concentric circle grey-scale fitting curve" feature provides a new perspective for quantifying the heterogeneity of internal structures within lesions, offering supplementary references for clinical early screening. Meanwhile, the results of multi-model comparisons provide experimental evidence for the selection and optimization of subsequent algorithms.
References
[1]. Zhou Yang, Zhou Xiu, Zhou Zhihui, et al. Application efficacy of artificial intelligence combined with ultrasound examination in distinguishing benign from malignant breast nodules [J]. Journal of Medical Imaging, 2025, 35(6): 169-172.
[2]. Gao Dixiao, Xia Huilin. Current Status and Prospects of Artificial Intelligence in Breast Cancer Ultrasound Diagnosis [J]. Journal of Inner Mongolia Medicine, 2025, 57(2): 175-178.
[3]. Pan Hao. Artificial Intelligence-Based Breast Cancer Auxiliary Diagnosis System [Doctoral Dissertation]. Xijing University, 2024.
[4]. Pons X, Marti-Botella N, Salcedo-Sanz S, et al. A maximum a posteriori framework for breast lesion segmentation in ultrasound images [J]. IEEE Transactions on Medical Imaging, 2014, 33(1): 214-225.
[5]. Huang X, Zhang Y, Li Z, et al. A framework for assisting breast cancer diagnosis using multi-modal breast ultrasound images [C]//International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2019: 552-560.
[6]. Ferrea R, Elst A J, Senthilnathan A, et al. Machine learning analysis of breast ultrasound texture features for the prediction of breast cancer subtypes [J]. Ultrasound in Medicine & Biology, 2022, 48(12): 2655-2664.
[7]. LEE Y W, HUANG C S, SHIH C C, et al. Axillary lymph node metastasis status prediction of early-stage breast cancer using convolutional neural networks [J]. Comput Biol Med, 2021, 130: 104206.
[8]. MOON W K, LEE Y W, KE H H, et al. Computer-aided diagnosis of breast ultrasound images using ensemble learning from convolutional neural networks [J]. Comput Methods Programs Biomed, 2020, 190: 105361.
[9]. Han Yang, Fu Dan, Bao Haihua, et al. Advances in deep learning for breast cancer ultrasound imaging [J]. Chinese Journal of Interventional Imaging and Therapy, 2025, 22(1): 71-74.
[10]. ZHANG X, ZHENG Y, YUAN Y, et al. Real-time breast lesion detection on ultrasound images via lightweight convolutional neural network [J]. IEEE Access, 2020, 8: 121078-121086.
[11]. GAO Siqi, Niu Si Hua, HUANG Jianhua, et al. Application of Ultrasound Artificial Intelligence in the Differential Diagnosis of Benign and Malignant Breast Diseases [J]. Chinese Journal of Ultrasound in Medicine, 2021, 37(7): 752-755.
[12]. Wu Xiaona, Geng Kelai, Dong Yinghui, et al. Application value of clinical-ultrasound models in predicting breast cancer risk [J]. Chinese Journal of Medical Computer Imaging, 2024, 30(6): 735-743.
[13]. Zhou Jianhua, Yu Jinhua, et al. Deep learning-based image-pathology integration using preoperative ultrasound and biopsy histopathology images distinguishes early-stage ductal and non-ductal breast cancers [J]. eBioMedicine, 2023.
[14]. Chen Rui, Niu Xiaoyue, Liu Xue, et al. Application Value of Artificial Intelligence-Assisted Diagnostic Systems in Evaluating Breast BI-RADS Category 4 Nodules [J]. Chinese Journal of Clinical Medical Imaging, 2024, 35(07): 481-484+490.
[15]. Shen Jie, Liu Yajing, Mo Miao, et al. A study on the effectiveness of artificial intelligence-assisted ultrasound in identifying breast lesions in Chinese women [J]. Chinese Journal of Cancer, 2023, 33(11): 1002-1008. DOI: 10.19401/j.cnki.1007-3639.2023.11.005.
[16]. Li Chengsheng, Bao Qihan, Hao Xiaoyan, et al. Construction of a Postoperative Predictive Model for Pancreatic Cancer Based on the Random Forest Algorithm [J]. Journal of Jilin University (Medical Edition), 2022, 48(2): 426–432.
[17]. Prokhorenkova L, Gusev G, Vorobev A, et al. CatBoost: unbiased boosting with categorical features [C]//Proceedings of the 32nd International Conference on Neural Information Processing Systems. 2018: 6890–6901.
[18]. Aryan Sai Boddu, Aatifa Jan. A systematic review of machine learning algorithms for breast cancer detection. Tissue and Cell. 2025; 95: 102929. ISSN 0040-8166.
[19]. Rautela K, Kumar D, Kumar V. Active contour and texture features hybrid model for breast cancer detection from ultrasonic images. International Journal of Imaging Systems and Technology. 2023; 33(6): 2061-2072. doi: 10.1002/ima.22909
Cite this article
Zeng,H. (2025). Analysis of the Effectiveness of the Slope of the Concentric Circle Grey-Scale Fitting Curve in Breast Cancer Ultrasound Diagnosis. Theoretical and Natural Science,141,1-10.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of ICBioMed 2025 Symposium: AI for Healthcare: Advanced Medical Data Analytics and Smart Rehabilitation
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Zhou Yang, Zhou Xiu, Zhou Zhihui, et al. Application efficacy of artificial intelligence combined with ultrasound examination in distinguishing benign from malignant breast nodules [J]. Journal of Medical Imaging, 2025, 35(6): 169-172.
[2]. Gao Dixiao, Xia Huilin. Current Status and Prospects of Artificial Intelligence in Breast Cancer Ultrasound Diagnosis [J]. Journal of Inner Mongolia Medicine, 2025, 57(2): 175-178.
[3]. Pan Hao. Artificial Intelligence-Based Breast Cancer Auxiliary Diagnosis System [Doctoral Dissertation]. Xijing University, 2024.
[4]. Pons X, Marti-Botella N, Salcedo-Sanz S, et al. A maximum a posteriori framework for breast lesion segmentation in ultrasound images [J]. IEEE Transactions on Medical Imaging, 2014, 33(1): 214-225.
[5]. Huang X, Zhang Y, Li Z, et al. A framework for assisting breast cancer diagnosis using multi-modal breast ultrasound images [C]//International Conference on Medical Image Computing and Computer-Assisted Intervention. Springer, Cham, 2019: 552-560.
[6]. Ferrea R, Elst A J, Senthilnathan A, et al. Machine learning analysis of breast ultrasound texture features for the prediction of breast cancer subtypes [J]. Ultrasound in Medicine & Biology, 2022, 48(12): 2655-2664.
[7]. LEE Y W, HUANG C S, SHIH C C, et al. Axillary lymph node metastasis status prediction of early-stage breast cancer using convolutional neural networks [J]. Comput Biol Med, 2021, 130: 104206.
[8]. MOON W K, LEE Y W, KE H H, et al. Computer-aided diagnosis of breast ultrasound images using ensemble learning from convolutional neural networks [J]. Comput Methods Programs Biomed, 2020, 190: 105361.
[9]. Han Yang, Fu Dan, Bao Haihua, et al. Advances in deep learning for breast cancer ultrasound imaging [J]. Chinese Journal of Interventional Imaging and Therapy, 2025, 22(1): 71-74.
[10]. ZHANG X, ZHENG Y, YUAN Y, et al. Real-time breast lesion detection on ultrasound images via lightweight convolutional neural network [J]. IEEE Access, 2020, 8: 121078-121086.
[11]. GAO Siqi, Niu Si Hua, HUANG Jianhua, et al. Application of Ultrasound Artificial Intelligence in the Differential Diagnosis of Benign and Malignant Breast Diseases [J]. Chinese Journal of Ultrasound in Medicine, 2021, 37(7): 752-755.
[12]. Wu Xiaona, Geng Kelai, Dong Yinghui, et al. Application value of clinical-ultrasound models in predicting breast cancer risk [J]. Chinese Journal of Medical Computer Imaging, 2024, 30(6): 735-743.
[13]. Zhou Jianhua, Yu Jinhua, et al. Deep learning-based image-pathology integration using preoperative ultrasound and biopsy histopathology images distinguishes early-stage ductal and non-ductal breast cancers [J]. eBioMedicine, 2023.
[14]. Chen Rui, Niu Xiaoyue, Liu Xue, et al. Application Value of Artificial Intelligence-Assisted Diagnostic Systems in Evaluating Breast BI-RADS Category 4 Nodules [J]. Chinese Journal of Clinical Medical Imaging, 2024, 35(07): 481-484+490.
[15]. Shen Jie, Liu Yajing, Mo Miao, et al. A study on the effectiveness of artificial intelligence-assisted ultrasound in identifying breast lesions in Chinese women [J]. Chinese Journal of Cancer, 2023, 33(11): 1002-1008. DOI: 10.19401/j.cnki.1007-3639.2023.11.005.
[16]. Li Chengsheng, Bao Qihan, Hao Xiaoyan, et al. Construction of a Postoperative Predictive Model for Pancreatic Cancer Based on the Random Forest Algorithm [J]. Journal of Jilin University (Medical Edition), 2022, 48(2): 426–432.
[17]. Prokhorenkova L, Gusev G, Vorobev A, et al. CatBoost: unbiased boosting with categorical features [C]//Proceedings of the 32nd International Conference on Neural Information Processing Systems. 2018: 6890–6901.
[18]. Aryan Sai Boddu, Aatifa Jan. A systematic review of machine learning algorithms for breast cancer detection. Tissue and Cell. 2025; 95: 102929. ISSN 0040-8166.
[19]. Rautela K, Kumar D, Kumar V. Active contour and texture features hybrid model for breast cancer detection from ultrasonic images. International Journal of Imaging Systems and Technology. 2023; 33(6): 2061-2072. doi: 10.1002/ima.22909









