1. Introduction
The lung is the main organ of the respiratory system, and X-ray examination provides precise information about lung shadows, lesion morphology, and other properties. With the outbreak of Covid-19, there has been an increasing focus on lung health, making lung health monitoring more prevalent. Traditional image analysis methods use manual segmentation and statistical classification to identify lung infections, but these methods are inefficient. In recent years, the development of convolutional neural networks (CNNs) has significantly improved the ability to classify and detect objects in image analysis.
However, in the pursuit of accuracy, many studies have overlooked the lightweight aspect of models. In previous research, some models have reached depths of hundreds of layers or more, resulting in slower model execution, longer monitoring times per instance, reduced monitoring efficiency, and increased monitoring costs. These factors limit the practical application of these models in monitoring scenarios. This study aims to investigate and compare the efficiency and accuracy of three CNN models, namely VGG16, MobileNetV2, and InceptionV3, in lung infection detection tasks.
Considering the limited number of training samples in the original dataset, we employed geometric data augmentation techniques to expand the dataset and improve the training effectiveness. In this study, we selected VGG16, MobileNetV2, and InceptionV3, which are well-established CNN models in image classification, known for their excellent performance and reliability, for comparison. By evaluating the lightweight nature and accuracy of these models, we will be able to determine the most suitable model for lung infection detection tasks.
This research holds significant importance in achieving lightweight models for lung infection detection. By reducing model complexity and parameter count, it becomes possible to lower the cost of medical devices and enhance the feasibility of deploying the models in practical applications, including remote healthcare scenarios. Furthermore, this study provides a foundation for further improvements and optimizations in lung infection detection technology, thereby enhancing the efficiency and accuracy of medical diagnosis and monitoring.
2. Related Work
Data augmentation methods have been widely used in deep learning tasks, but they require manual design. For natural image datasets like CIFAR-10 and ImageNet, common methods include image translations and horizontal reflections [1]. In this study, we performed data augmentation operations such as translation, rotation, and scaling on the Chest X-Ray Images dataset.
Deep learning has been extensively applied to classification problems. In the mentioned studies, the Vgg16 model was employed for facial expression recognition tasks in the education domain [2], while the MobileNetV2 model was used for fruit image classification [3]. Additionally, Qian compared the performance of models such as InceptionV3 and Vgg16 in galaxy morphology classification [4].
In the field of medical imaging, deep learning has shown remarkable capabilities for disease detection and classification tasks. Rahman et al. utilized AlexNet, ResNet18, DenseNet201, and SqueezeNet for the detection of three types of pneumonia (normal, bacterial pneumonia, and viral pneumonia) using a total of 5247 images. Among them, the DenseNet201 model achieved the best prediction accuracy of 98% [5]. However, due to its depth of 201 layers, the DenseNet201 model has a large number of parameters, making it prone to overfitting. Furthermore, Kim et al. applied the EfficientNet V2-M deep learning model to the multiclass classification of chest X-ray images for various lung diseases, with a total of 10,000 images for normal, pneumonia, and pneumothorax. They achieved validation performance with an accuracy of 82.15%, sensitivity of 81.40%, and specificity of 91.65%. However, this model has a large number of parameters, reaching 53,155,512, requiring further model compression [6]. Ayan et al. compared the performance of Vgg16 and Xception models on the Chest X-Ray Images dataset, with the best recall rate and accuracy being 89.1% and 84.5%, respectively. The lower accuracy may be attributed to the small dataset size [7]. Apostolopoulos et al. demonstrated that MobileNetV2 outperforms VGG19 in terms of specificity on a dataset of only 1427 X-ray images [8]. However, due to the small dataset size, overfitting is likely to occur, and further validation should be performed after data augmentation.
These studies demonstrate the effectiveness of different deep learning models in medical image analysis tasks, including pneumonia detection and lung infection classification. However, further exploration of parameter compression techniques is still needed to improve the efficiency and practicality of these models in real-world applications, while also addressing the issue of overfitting in research.
3. Dataset and Geometric Transformation-based Data Augmentation
3.1. Chest X-Ray Images Dataset
Our study utilized the publicly available Chest X-Ray Pneumonia database sourced from Kaggle, which originated from Mendeley Data [9]. This database consists of three folders: training, testing, and validation sets, each containing subfolders representing two classes (Pneumonia/Normal). A total of 5,856 X-ray images in JPEG format were included in the dataset, distributed across the Pneumonia and Normal categories. The training set comprised 5,216 samples, with Pneumonia samples accounting for 74.29% of the total; the testing set consisted of 634 samples, with Pneumonia samples representing 62.50% of the total; and the validation set contained 16 samples, with Pneumonia samples comprising 50% of the total. Pneumonia samples could be attributed to viral infection, bacterial infection, or mixed infection. Figure 1 illustrates six randomly selected image samples from different categories in the dataset, wherein images (a), (b), and (c) depict healthy lung structures, while images (d), (e), and (f) exhibit typical radiographic features of pneumonia, such as infiltrations and shadows. By conducting in-depth examination and analysis of these images, exploring the morphological characteristics of pneumonia lesions in comparison to normal lung structures, we can gain further insights into the diverse manifestations of pneumonia, enhance early detection rates, alleviate the burden on the healthcare system, and improve treatment outcomes for patients.
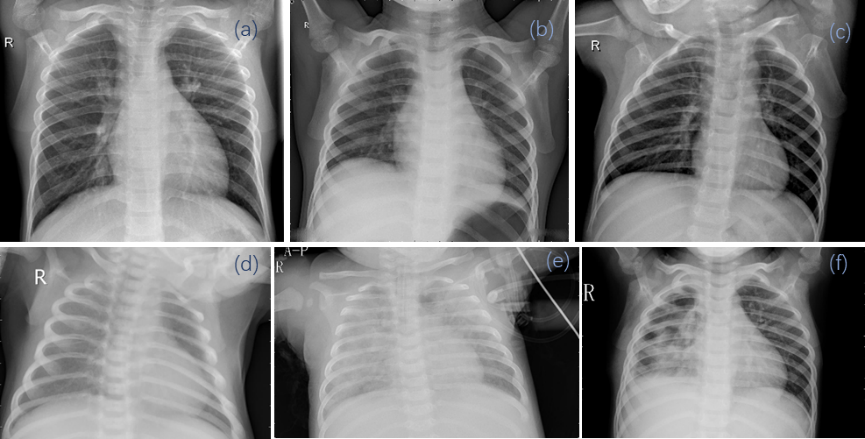
Figure 1. Original data in the dataset: Images of normal (a, b, c) and pneumonia (d, e, f) cases
3.2. Data Preprocessing and Augmentation
Each image must undergo preprocessing based on the specific deep neural network being used, involving resizing and normalization. Different neural networks require images of different sizes based on their architecture. MobileNetV2 requires images of size 224 × 224, while InceptionV3 and VGG16 require images of size 229 × 229.
Training a neural network requires a large amount of data. In real-world scenarios, doctors can use different operations such as rotation on various images. However, the original data is not robust enough as they have similar rotations, brightness, etc., which can deteriorate the generalization capability of the neural network. Data augmentation effectively utilizes existing data to enhance the neural network's ability to handle complex images and reduces the occurrence of model overfitting. In this study, the Chest X-Ray Healthy Image dataset was subjected to various augmentation techniques using the ImageDataGenerator in TensorFlow. These operations diversify the images in terms of position, orientation, brightness, and other aspects, providing a more diverse training dataset, thus improving the performance and robustness of the model. The following augmentation operations were performed on the Chest X-Ray Healthy Image dataset: 1) Image rotation: Randomly rotate the image by 0-10 degrees, providing spatial variation in position and orientation. 2) Translation: Perform horizontal and vertical shifts on the image, with shift distances set to 10% of the image size. This introduces minor variations in position, increasing data diversity. 3) Shearing: Apply shearing transformation to the image, shearing it counterclockwise by 0.5. This introduces distortion in the image's orientation. 4) Scaling: Simultaneously scale the length and width of the image by the same factor, altering the image's size. 5) Brightness adjustment: Randomly modify the image's brightness during the data augmentation process. This simulates changes in lighting conditions and increases data diversity.
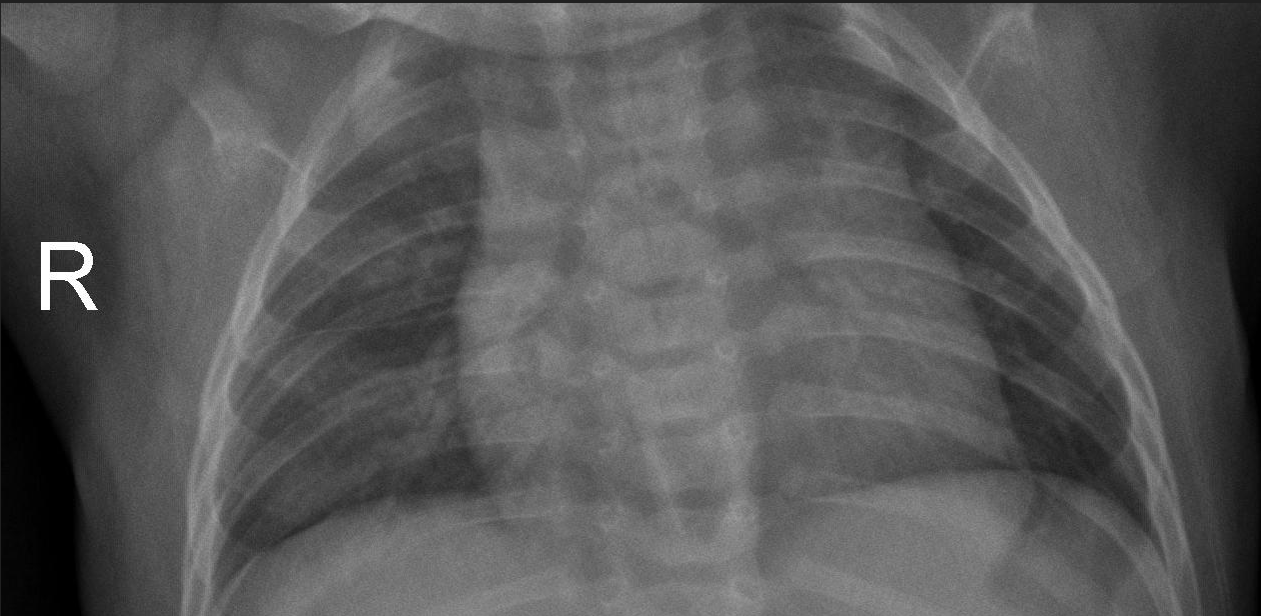
Figure 2. Comparison of Data Augmentation: Original images before augmentation (Top 1) and augmented images after data augmentation (Bottom 4).
As illustrated in Figure 2, data augmentation enhances the model's generalization capability when trained on the dataset, while also mitigating overfitting on small samples, thereby facilitating the comparison of performance metrics for subsequent models.
4. VGG16, MobileNetV2, and InceptionV3
4.1. Three Models
The InceptionV3 model is the third-generation model in Google's Inception series, which introduces several innovative ideas in neural network design [10]. In comparison to other neural network models, the most notable characteristic of the Inception network is the expansion of convolutional operations between network layers. It utilizes convolutional kernels of different sizes to obtain receptive fields of varying scales and combines features from different scales through concatenation.
The core of InceptionV3 is represented as follows: Here, X represents the input feature map, Conv denotes convolutional operation, MaxPooling represents max pooling operation, and Concat indicates feature concatenation operation. By employing multiple convolutional kernels and MaxPooling at different scales, the InceptionV3 model can simultaneously capture features at various scales and concatenate them to enrich the model's representation of image features. The formula is expressed as follows:
\( InceptionV3(X)=Concat([Conv(X),Conv(X),Conv(X),MaxPooling(X)])\ \ \ (1) \)
VGG (Visual Geometry Group), proposed by the Visual Geometry Group at Oxford [11], has made a significant contribution by demonstrating that increasing the depth of a network can improve its performance to a certain extent. The VGG network has two variations: VGG16 and VGG19, with the main difference lying in their depths. In this study, we selected the VGG16 model, which is suitable for the Kaggle Chest X-Ray Healthy Image dataset.
The MobileNetV2 network is an improved version proposed by the Google team in 2018. Compared to the MobileNetV1 network, MobileNetV2 exhibits improvements in both accuracy and model size. It adopts a structure called "Inverted Residuals with Linear Bottlenecks" [12], which utilizes depthwise separable convolutions and linear bottlenecks to achieve the design of a lightweight model.
The architecture of the MobileNetV2 model is represented as follows: Here, X represents the input feature map, Conv denotes the convolutional operation, DepthwiseConv represents the depthwise separable convolution operation, and Bottleneck represents the linear bottleneck connection. MobileNetV2 reduces the computational complexity through depthwise separable convolutions and ensures rich feature representation through linear bottleneck connections. This design allows MobileNetV2 to maintain high accuracy while being lightweight, making it suitable for deployment in resource-constrained environments. The formula is expressed as follows:
\( MobileNetV2(X)=Bottleneck(DepthwiseConv(Conv(X)))\ \ \ (2) \)
Among the three models mentioned above, VGG16 adopts the classic structure of stacked convolutional layers and fully connected layers. MobileNetV2 reduces the number of parameters and computational complexity through depthwise separable convolutions. InceptionV3 introduces Inception modules that are stacked repeatedly to form a larger network, enhancing feature extraction capabilities. In this study, we utilized InceptionV3, VGG16, MobileNetV2 [10, 11, 12] as the base models to evaluate their performance on the classification task using the Chest X-Ray Images (Pneumonia) dataset.
4.2. Training Configuration
The code in the article is written in Python 3.11 and Tensorflow 2.15. We performed classification tasks on both the training and testing datasets. Take the operations on MobileNetV2 as an example which is shown in Tables 1. For InceptionV3, we input the original data and augmented data (images are all 229*229*3) into the InceptionV3 model, resulting in an output of 5*5*2048. After applying dropout, flattening, full connection operation, another dropout, and two additional full connection operations, each image is processed into a 1*256 tensor. For VGG16, the operations are similar to InceptionV3. For MobileNetV2, we incorporated a 2D global average pooling operation before the first dropout, which served as a structural regularization to prevent overfitting for the entire network.
Table 1. MobileNetV2
Layer | Out Shape | Param |
input_layer | (None,224, 224,3) | 0 |
augmentation_layer | (None, None, 3) | 0 |
mobilenetv2 (Functional) | (None, 7, 7, 1280) | 2257984 |
global_average_pooling2d | (None, 1280) | 0 |
dropout_1 (Dropout) | (None, 1280) | 0 |
flatten (Flatten) | (None, 1280) | 0 |
dense_1 (Dense) | (None, 512) | 655872 |
dropout_2 (Dropout) | (None, 512) | 0 |
dense_2 (Dense) | (None, 256) | 131328 |
dense_3 (Dense) | (None, 1) | 257 |
5. Results
5.1. Evaluation Metrics and Evaluation Methods
After the completion of training, all models were tested on the testing dataset. The performance of each model was evaluated using metrics such as accuracy, recall, precision, F1-Score, and the area under the ROC curve (AUC). All models showed convergence of the loss function during training, as exemplified by the InceptionV3 model without data augmentation in Figure 3.
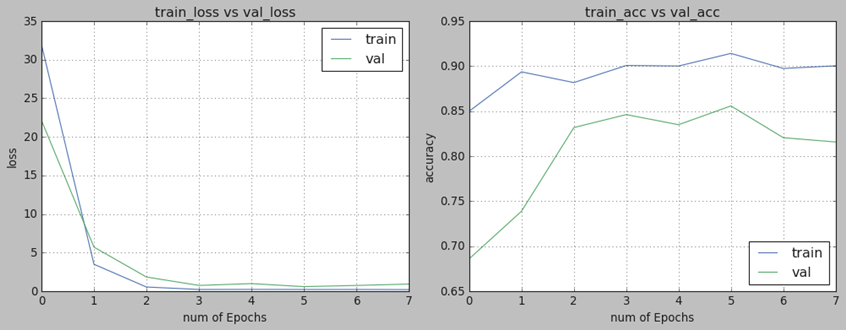
Figure 3. Training Progress of InceptionV3
Next, we will discuss the performance metrics used in the paper. As shown in Figure 4, there are 6 confusion matrixes, which are in standardized format for evaluating image classification accuracy. The first row represents the confusion matrix without data augmentation, while the second row represents the confusion matrix with data augmentation. It can be observed that the diagonal elements of the confusion matrix with data augmentation have relatively darker colors, indicating a higher classification accuracy of the models with data augmentation.
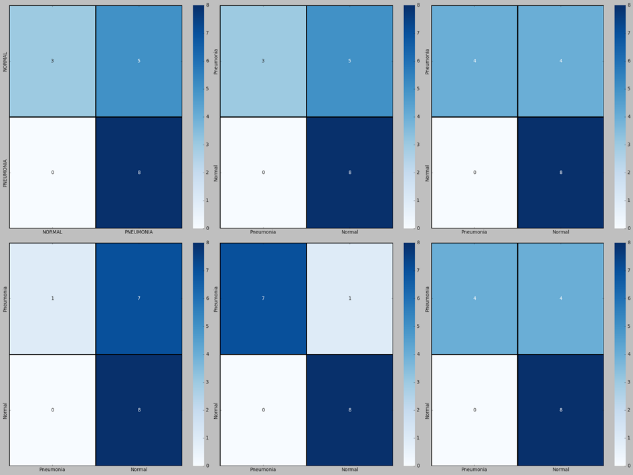
Figure 4. From left to right: InceptionV3, VGG16, MobileNetV2 models
5.2. Model Comparison and Analysis
Table 2 below presents the parameter usage and performance scores of each model. We compared InceptionV3, VGG16, and MobileNetV2 in terms of F1-Score, average precision, average accuracy, and the area under the ROC curve before and after data augmentation. The results demonstrate that these metrics have improved after data augmentation, highlighting the superiority of data augmentation in enhancing model training accuracy. Specifically, the VGG16 - Augmentation model achieved the highest average accuracy of 96.8%. The MobileNetV2 model, after data augmentation, also reached an average accuracy of 94.2%. It is noteworthy that the MobileNetV2 model has the fewest parameters, only 1/9 of the parameters of the VGG16 - Augmentation model, indicating its higher model lightweight performance.
Overall, through data augmentation, models such as InceptionV3, VGG16, and MobileNetV2 showed improvements in F1-Score, average precision, average accuracy, and the area under the ROC curve. The VGG16 - Augmentation model achieved the highest average accuracy, while the MobileNetV2 - Augmentation model demonstrated relatively high average accuracy with the smallest parameter count, highlighting its advantage in lightweight modeling. These results further validate the effectiveness and applicability of data augmentation in enhancing model accuracy.
Table 2. Performance Comparison of the Three Models Before (After) Augmentation
Model | Param | F1 - Score | ROC - AUC | Accuracy | Precision |
InceptionV3 | 48,149,281 | 0.6957 | 0.5625 | 0.908725 | 0.919975 |
VGG16 | 27,691,841 | 0.6957 | 0.5625 | 0.968283 | 0.979183 |
MobileNetV2 | 3,045,441 | 0.7619 | 0.6875 | 0.942580 | 0.961615 |
InceptionV3 with augmentation | 48,149,281 | 0.9412 | 0.9375 | 0.838780 | 0.834680 |
VGG16 with augmentation | 27,691,841 | 0.8000 | 0.7500 | 0.936675 | 0.959550 |
MobileNetV2 with augmentation | 3,045,441 | 0.8000 | 0.7500 | 0.913900 | 0.944314 |
6. Conclusions
This study compared the results of three models and found that VGG16 - Augmentation achieved the highest accuracy in the task of detecting lung infections (96.8%). However, in real-time monitoring scenarios where we need to monitor a large amount of healthy lung data, the parameter size of VGG16 - Augmentation would be excessively large, which is not conducive to efficient monitoring. On the other hand, MobileNetV2 - Augmentation, although having a moderate accuracy performance (94.2%), it only has 1/9 of the parameters of the VGG16 - Augmentation model, making it potentially more practical in real-time monitoring scenarios.
Nevertheless, it is acknowledged that there is room for improvement in our work. Firstly, in terms of data volume, the chest X-ray image dataset does not cover all possible scenarios. Therefore, larger-scale datasets are needed to further validate our models. Secondly, exploring higher-performing network architectures or more powerful data augmentation techniques is necessary to improve the classification performance of the models in terms of accuracy. Additionally, combining predictions from multiple models through ensemble learning methods can further enhance classification accuracy. Thirdly, to validate the robustness of the models, training and testing the MobileNetV2 model on different datasets can be conducted to assess the reliability and stability of the model under different data distributions. Fourthly, while our models have adopted lightweight architectures such as MobileNetV2, further optimization of model size and computational complexity can be pursued to meet the demands of real-time monitoring in clinical or remote healthcare settings.
In conclusion, although we have achieved certain results, there is still room for further improvement in terms of data validation, accuracy, lightweightness, and robustness. Validation of the models can be done by utilizing larger-scale datasets and training/testing on different datasets, while deeper research in model design and optimization can enhance the application value of our proposed image classification techniques in clinical practice.
References
[1]. A. Krizhevsky, I. Sutskever, and G. E. Hinton (2012). Imagenet classification with deep convolutional neural networks. Communications of the ACM, 60, 84–90. https://doi.org/10.1145/3065386
[2]. Lawpanom, R.; Songpan, W.; Kaewyotha, J. (2024). Advancing Facial Expression Recognition in Online Learning Education Using a Homogeneous Ensemble Convolutional Neural Network Approach. Applied Sciences, 14(3). https://doi.org/10.3390/app14031156
[3]. Gulzar, Y. (2023). Fruit Image Classification Model Based on MobileNetV2 with Deep Transfer Learning Technique. Sustainability, 15(3). https://doi.org/10.3390/su15031906
[4]. Qian. (2023). Performance comparison among VGG16, InceptionV3, and resnet on galaxy morphology classification. Journal of Physics, 2580. https://dx.doi.org/10.1088/1742-6596/2580/1/012009
[5]. Rahman, T.; Chowdhury, M.E.H.; Khandakar, A.; Islam, K.R.; Islam, K.F.; Mahbub, Z.B.; Kadir, M.A.; Kashem, S. (2020). Transfer Learning with Deep Convolutional Neural Network (CNN) for Pneumonia Detection Using Chest X-ray. Applied Sciences, 10(9). https://doi.org/10.3390/app10093233
[6]. Kim S, Rim B, Choi S, Lee A, Min S, Hong M. (2022). Deep Learning in Multi-Class Lung Diseases' Classification on Chest X-ray Images. Diagnostics (Basel), 12(4). https://doi.org/10.3390/diagnostics12040915
[7]. E. Ayan and H. M. Ünver (2019). Diagnosis of Pneumonia from Chest X-Ray Images Using Deep Learning. Scientific Meeting on Electrical-Electronics & Biomedical Engineering and Computer Science (EBBT), 1-5. https://ieeexplore.ieee.org/document/8741582
[8]. Apostolopoulos, I.D., Mpesiana, T.A. (2020). Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med, 43, 635–640. https://link.springer.com/article/10.1007/s13246-020-00865-4
[9]. Kermany, Daniel S., Kang Zhang and Michael H. Goldbaum(2018). Labeled Optical Coherence Tomography (OCT) and Chest X-Ray Images for Classification. https://api.semanticscholar.org/CorpusID:126183849
[10]. C. Szegedy, V. Vanhoucke, S. Ioffe, J. Shlens and Z. Wojna (2016). Rethinking the Inception Architecture for Computer Vision. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2818-2826. https://ieeexplore.ieee.org/document/7780677
[11]. Simonyan K, Zisserman A (2014). Very Deep Convolutional Networks for Large-Scale Image Recognition. CoRR, abs/1409.1556. https://doi.org/10.48550/arXiv.1409.1556
[12]. M. Sandler, A. Howard, M. Zhu, A. Zhmoginov and L. -C. Chen (2018). MobileNetV2: Inverted Residuals and Linear Bottlenecks. 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, 4510-4520. https://ieeexplore.ieee.org/abstract/document/8578572
Cite this article
Zhang,Y.;Xu,Y.;Kong,Z.;Hu,Z. (2024). Comparison of deep learning models based on Chest X-ray image classification. Applied and Computational Engineering,64,55-62.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 6th International Conference on Computing and Data Science
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. A. Krizhevsky, I. Sutskever, and G. E. Hinton (2012). Imagenet classification with deep convolutional neural networks. Communications of the ACM, 60, 84–90. https://doi.org/10.1145/3065386
[2]. Lawpanom, R.; Songpan, W.; Kaewyotha, J. (2024). Advancing Facial Expression Recognition in Online Learning Education Using a Homogeneous Ensemble Convolutional Neural Network Approach. Applied Sciences, 14(3). https://doi.org/10.3390/app14031156
[3]. Gulzar, Y. (2023). Fruit Image Classification Model Based on MobileNetV2 with Deep Transfer Learning Technique. Sustainability, 15(3). https://doi.org/10.3390/su15031906
[4]. Qian. (2023). Performance comparison among VGG16, InceptionV3, and resnet on galaxy morphology classification. Journal of Physics, 2580. https://dx.doi.org/10.1088/1742-6596/2580/1/012009
[5]. Rahman, T.; Chowdhury, M.E.H.; Khandakar, A.; Islam, K.R.; Islam, K.F.; Mahbub, Z.B.; Kadir, M.A.; Kashem, S. (2020). Transfer Learning with Deep Convolutional Neural Network (CNN) for Pneumonia Detection Using Chest X-ray. Applied Sciences, 10(9). https://doi.org/10.3390/app10093233
[6]. Kim S, Rim B, Choi S, Lee A, Min S, Hong M. (2022). Deep Learning in Multi-Class Lung Diseases' Classification on Chest X-ray Images. Diagnostics (Basel), 12(4). https://doi.org/10.3390/diagnostics12040915
[7]. E. Ayan and H. M. Ünver (2019). Diagnosis of Pneumonia from Chest X-Ray Images Using Deep Learning. Scientific Meeting on Electrical-Electronics & Biomedical Engineering and Computer Science (EBBT), 1-5. https://ieeexplore.ieee.org/document/8741582
[8]. Apostolopoulos, I.D., Mpesiana, T.A. (2020). Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Phys Eng Sci Med, 43, 635–640. https://link.springer.com/article/10.1007/s13246-020-00865-4
[9]. Kermany, Daniel S., Kang Zhang and Michael H. Goldbaum(2018). Labeled Optical Coherence Tomography (OCT) and Chest X-Ray Images for Classification. https://api.semanticscholar.org/CorpusID:126183849
[10]. C. Szegedy, V. Vanhoucke, S. Ioffe, J. Shlens and Z. Wojna (2016). Rethinking the Inception Architecture for Computer Vision. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2818-2826. https://ieeexplore.ieee.org/document/7780677
[11]. Simonyan K, Zisserman A (2014). Very Deep Convolutional Networks for Large-Scale Image Recognition. CoRR, abs/1409.1556. https://doi.org/10.48550/arXiv.1409.1556
[12]. M. Sandler, A. Howard, M. Zhu, A. Zhmoginov and L. -C. Chen (2018). MobileNetV2: Inverted Residuals and Linear Bottlenecks. 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, 4510-4520. https://ieeexplore.ieee.org/abstract/document/8578572









