1. Introduction
A Large Language Model (LLM) is a type of artificial intelligence designed to process and generate human-like text based on patterns learned from extensive datasets. These models, which can have tens to hundreds of billions of parameters, are trained on large volumes of textual content[1][2]. Utilizing deep learning techniques, particularly transformer architectures, LLMs are capable of understanding context and generating responses across a wide range of domains[3]. In the medical field, LLMs hold great promise, with potential applications including the generation of clinical documents, the provision of initial diagnostic recommendations, and the delivery of personalized medical advice[4]. However, the closed-source nature of models like GPT-4 and ChatGPT restricts their customization for specific medical needs. Additionally, these models may generate "hallucinations"—confident yet factually incorrect responses—often due to insufficient training on specialized medical datasets[5].
To address these issues, specialized medical LLMs, such as Google Research’s Med-PaLM[6], have demonstrated the ability to achieve human expert-level performance in answering US Medical Licensing Examination (USMLE)-style questions[7]. However, Med-PaLM remains a closed-source model, with undisclosed details about its training data and architecture. Conversely, open-source models offer opportunities for customization and development tailored to specific medical contexts. For instance, Qilin-Med-7B, based on the Baichuan-7B open-source large model, focuses on Chinese medical knowledge[8]. However, its regional focus limits its applicability on a global scale[9].
To overcome these limitations, we have developed a new medical domain large model, “Cura-LLaMA,” based on the open-source TinyLlama model, which shares the same architecture and tokenizer as LLaMA2[10]. TinyLlama is a compact, open-source large language model, with a size of under 640 megabytes and 1.1 billion parameters. Its compactness offers significant advantages in reducing computational costs and memory usage, making it suitable for deployment on standard computers[11]. This feature is particularly beneficial in regions with limited access to high-performance computing resources, such as rural clinics or developing countries, thereby broadening the accessibility of medical AI support. Additionally, TinyLlama's efficiency potentially leads to faster response times.
The development of Cura-LLaMA involved several key stages. First, we identified the data necessary for training our model—PubMedQA, a specialized dataset designed for medical question-answering[12]. PubMedQA includes 1,000 expert-labeled instances, 61,200 unlabeled instances, and 211,300 artificially generated QA instances[13]. We then integrated these extensive PubMedQA datasets into TinyLlama to create Cura-LLaMA. By tailoring TinyLlama with this medical-specific data, we enhanced its ability to handle complex medical queries and generate reliable, contextually appropriate responses. Finally, we released Cura-LLaMA and compared its performance with other open-source specialized medical models currently available. Our goal is to make Cura-LLaMA a practical and accessible tool for the global medical community.
2. Related Work
2.1. Large Language Models
The foundational model for developing Cura-LLaMA, TinyLlama, is based on the architecture and tokenizer of LLaMA2[14]. LLaMA2 encompasses a series of LLMs with parameter counts ranging from 7 billion to 70 billion[15], pretrained on publicly available online data sources with 2 trillion tokens[16]. Another noteworthy open-source large language model is Mistral-7B[17], an LLM with 7.3 billion parameters, featuring advanced attention mechanisms such as grouped-query attention (GQA) and sliding window attention (SWA). GQA enhances the model’s focus and response relevance, while SWA optimizes attention within specific text windows, allowing for efficient management of long text sequences, thereby improving processing speed and reducing computational demands[18].
2.2. Fine-Tuning
Large Language Models (LLMs) are fine-tuned by training on task-specific datasets following their initial pre-training on broad datasets, adapting these pre-trained models to perform specialized tasks or meet specific use case requirements. Fine-tuning not only enhances the model’s performance on specialized tasks but also mitigates issues like overfitting, where a model trained on a limited dataset might perform well on that data but poorly on unseen data. Fine-tuning is generally more feasible and less resource-intensive than training a new model from scratch for a particular purpose[19]. Vicuna-13B is an example of how fine-tuning can significantly enhance a model’s performance. Starting with a foundational LLaMA model pretrained on diverse datasets, the Vicuna team fine-tuned it specifically for chatbot functions using 70K user-shared conversations from ShareGPT. In preliminary evaluations, Vicuna-13B demonstrated a relative response quality of 92%, significantly surpassing LLaMA-13B’s 68%, highlighting the effectiveness of fine-tuning in enhancing LLM capabilities[20].
2.3. Medical Domain LLMs
Several studies have developed fine-tuned, medical-specialized LLMs based on open-source pre-trained LLMs. For instance, PMC-LLaMA focuses on adapting the LLaMA model to the medical domain by incorporating domain-specific knowledge, including medical QA, rationale, and conversational dialogues, encompassing a total of 202 million tokens. Fine-tuning techniques were used to align the model with medical terminologies and scenarios. Evaluations of PMC-LLaMA against several medical QA benchmarks demonstrated its superior performance, even outperforming ChatGPT with 175B parameters in certain aspects, despite having only 13B parameters[21]. Another model, Me-LLaMA, utilizes a large-scale dataset consisting of 129 billion tokens to adapt the LLaMA2 model for medical contexts. It is claimed that Me-LLaMA outperforms other open-source medical LLMs in zero-shot, few-shot, and supervised learning scenarios across various datasets[22]. However, both models have limitations in generating responses that are not always factually accurate, as the veracity of all input data cannot be guaranteed.
3. Methodology
Evaluating TinyLlama: To evaluate the performance of the original TinyLlama model in the medical domain, we tested the chat model version, TinyLlama-1.1B-Chat-V0.4. This version was trained on datasets such as SlimPajama and the data used to train StarCoder[23]. SlimPajama is derived from the RedPajama dataset[24], which, according to Together AI, comprises seven data slices, none of which specifically target the medical domain[25]. The StarCoder dataset primarily incorporates over 80 different programming languages, as well as text extracted from GitHub issues, commits, and notebooks[26]. Consequently, the TinyLlama chat model was not explicitly designed for the medical field. Through our experiments, we aimed to elucidate the potential limitations and shortcomings of a general-purpose model when applied to specialized medical queries.
The experiments were conducted on an Alibaba Cloud gn6v server, equipped with an NVIDIA Tesla V100 GPU with 32 GB VRAM and a 12-core Intel Xeon Platinum 8163 processor, running Ubuntu 22.04 64-bit operating system. Model inference was performed using PyTorch 2.3.0+cu121 and Transformers 4.41.2, with additional support from libraries such as Numpy 1.26.4. All experiments were executed in this environment.
In this experiment, we utilized a subset of the PubMedQA dataset, specifically the pqa_labeled subset, for testing purposes. This subset comprises 1,000 expert-annotated question-answer (QA) instances, with each instance containing five components: pubid, question, context, long_answer, and final_decision. Due to constraints in cost and time, we limited our testing to the first 500 rows of this subset.
The testing methodology, Query-Response Consistency Analysis, was as follows: the medical questions in the [question] component were input into the TinyLlama chat model, and the model’s predicted outputs were compared against the corresponding final_decision. The final_decision can take one of three possible outcomes: yes, no, or maybe. We employed keyword detection for comparison: if the TinyLlama output contained any of the keywords yes, no, or maybe (with “yes and no” also classified as maybe), we compared it to the final_decision. If the two matched, the prediction was marked as correct; otherwise, it was marked as incorrect. If the model’s output did not contain any of the keywords, or if it contained two or more of the keywords yes, no, or maybe, it was categorized as “unable to locate keyword.”
It is important to note that the model occasionally generated irrelevant or nonsensical text (referred to as Garbage Text or Gibberish). For instance, in response to a medical question regarding laryngeal mask airway procedures, the model might output content related to TensorFlow or the implementation of Open Assistant projects, which are entirely unrelated to the medical query. Consequently, in this study, if the model’s response included any form of unintelligible or random text, such as coding errors, incoherent content, or unrelated technical references, it was considered an abnormal response, and the prediction was marked as incorrect.
The following code snippet illustrates the process of loading the dataset, generating predictions, and recording the results using the TinyLlama-1.1B-Chat-V0.4 model:
from datasets import load_dataset
from transformers import AutoTokenizer
import transformers
import torch
model = "TinyLlama/TinyLlama-1.1B-Chat-v0.4"
def inference(question):
pipeline = transformers.pipeline(
"text-generation",
model=model,
torch_dtype=torch.float16,
device_map="auto",
)
CHAT_EOS_TOKEN_ID = 32002
prompt = question
formatted_prompt = (
f"user\n{prompt}\nassistant\n"
)
sequences = pipeline(
formatted_prompt,
do_sample=True,
top_k=50,
top_p=0.9,
num_return_sequences=1,
repetition_penalty=1.1,
max_new_tokens=1024,
eos_token_id=CHAT_EOS_TOKEN_ID,
)
seq = sequences[0]
ret = seq['generated_text']
print(f"Result: {ret}")
return ret
data = load_dataset("qiaojin/pubmed_qa", 'pqa_labeled')
data = data['train']
NUM_SAMPLES = 496
for idx, sample in enumerate(data):
question = sample["question"]
final_decision = sample["final_decision"]
print(f'### Question: {question}\n### Final Decision: {final_decision}')
model_prediction = inference(sample["question"])
print(f'### Model Prediction: {model_prediction}\n\n')
with open('tinyllama_output.txt', 'a') as f:
f.write(f'### Question: {question}\n### Final Decision: {final_decision}\n### Model Prediction: {model_prediction}\n\n')
if idx == NUM_SAMPLES:
break
Fine-tuning for Cura-Llama: I employed the same computational setup described previously (see Evaluating TinyLlama section), leveraging the qiaojin/PubMedQA dataset available on Hugging Face for fine-tuning. To ensure that the test data did not overlap with the training data (pqa_labeled split), I selected the pqa_artificial subset, which consists of QA instances that have been meticulously annotated by experts, ensuring higher data quality. For this fine-tuning process, I utilized the first 1,000 rows of QA instances from the training split. To optimize the model’s performance under limited resources, I streamlined the dataset by retaining only the essential [question] and [final_decision] columns, discarding all other non-essential columns. Special tokens <im_start> and <im_end> are added to clearly delineate the question and the decision. This helps the model distinguish between input and output segments, improving its ability to generate accurate responses during fine-tuning.
The following code snippet demonstrates how the pqa_labeled subset of the PubMedQA dataset was processed to prepare it for fine-tuning the Cura-LLaMA model:
pubmed_dataset = pubmed_dataset.map(
lambda questiontext: {
"text": f"<im_start>{questiontext['text']}<im_end> <im_start>{questiontext['final_decision']}<im_end>"
})
During training, the number of epochs was set to 3, and the learning rate was fixed at 1e-5. This is aimed to ensure ensures that the model goes through a complete training cycle on all data but minimizing the risk of overfitting. I utilized Weights & Biases (wandb) to log and monitor the training process. The following outlines the key metrics and observations recorded during training:
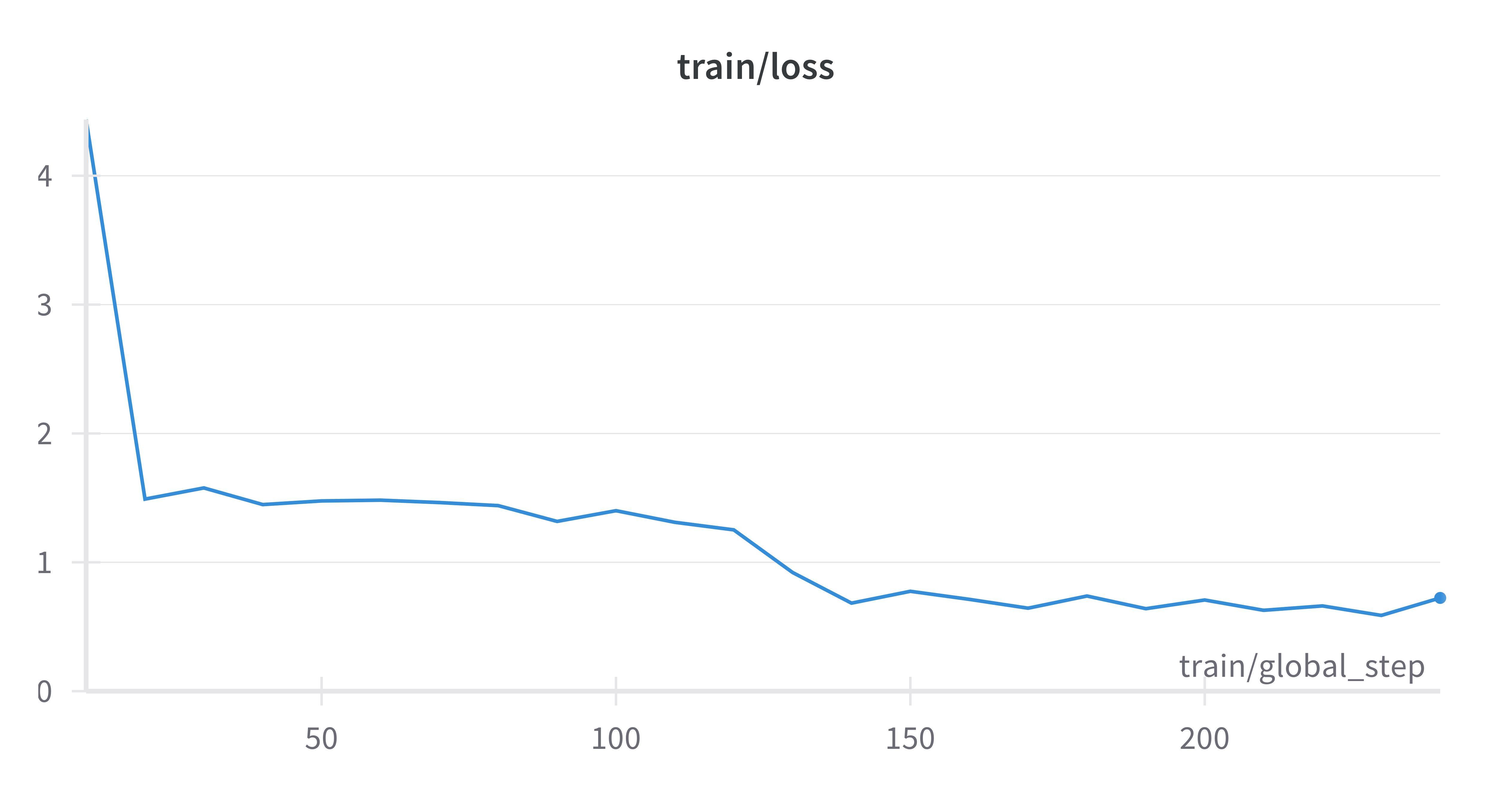
Figure 1. Training Loss Curve for Cura-LLaMA.
This is the train loss curve during the fine-tuning process, which shows expected behavior in a well-structured training process. The sharp initial drop in loss indicates that the model is quickly learning the basic patterns in the data. As training continues, the curve flattens, showing that the model is making finer adjustments to optimize its performance.
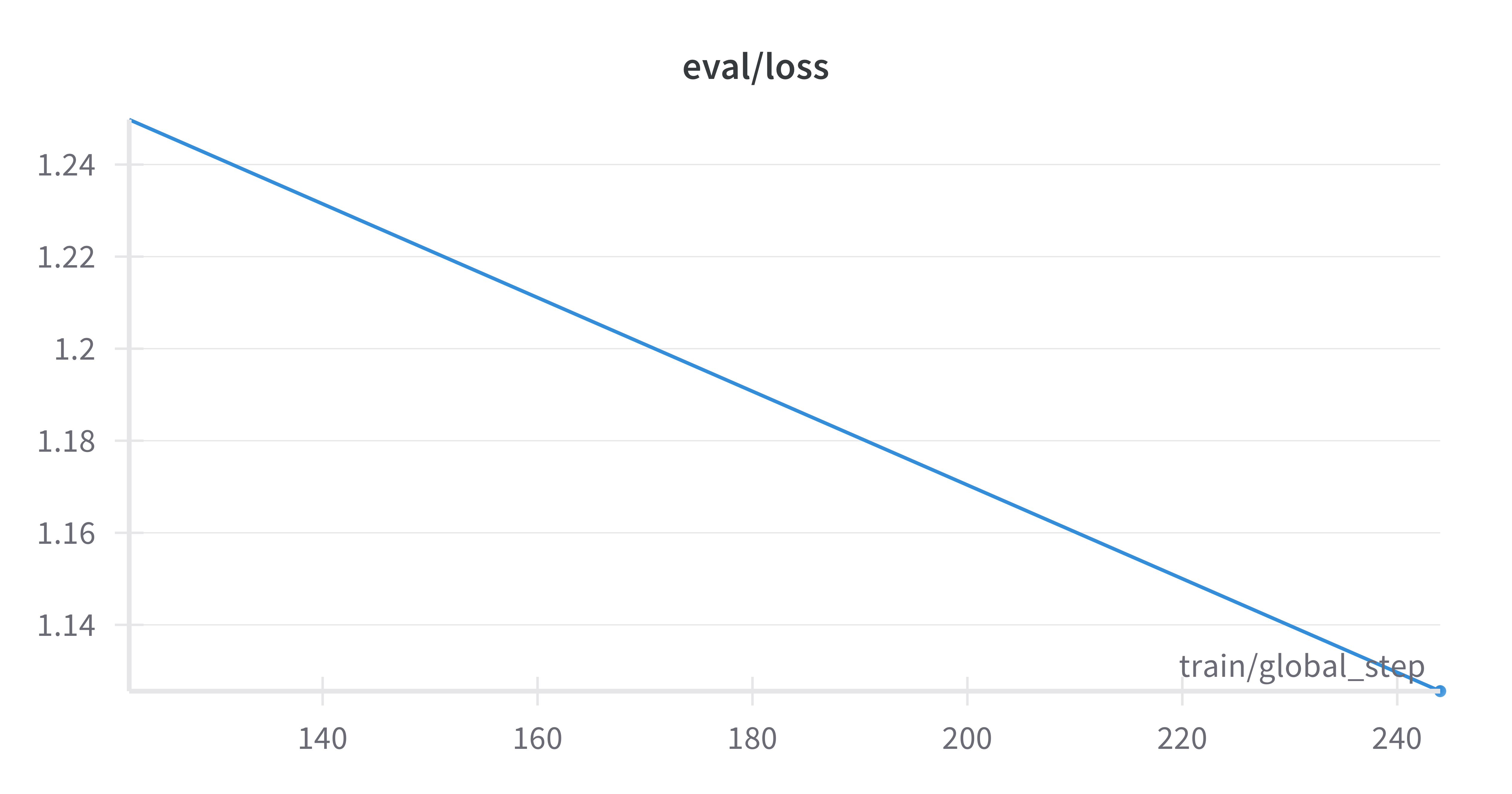
Figure 2. Evaluation Loss Curve for Cura-LLaMA
This is the evaluation loss curve during the fine-tuning process, indicating that the model is learning effectively without significant overfitting and improving its generalization capabilities.
To ensure consistency in our experiments and control for variables, we tested the fine-tuned model using the same dataset and methodology as described previously. We utilized the first 500 rows of the pqa_labeled subset for testing. The model’s performance in the medical domain was analyzed using the Keyword Detection Method (Query-Response Consistency Analysis).
4. Results
TinyLlama Chat Model: The performance of the TinyLlama Chat Model in predicting responses to nearly 500 PubMedQA questions was evaluated using the previously described keyword detection method, Query-Response Consistency Analysis. Out of the 496 prediction results analyzed, 16 were identified as containing "Garbage Text or Gibberish." These abnormal responses predominantly included content related to machine learning algorithms, data analysis, cloud computing, and other computer science fields, thus failing to address the medical questions posed. Regardless of whether the response matched the [final_decision] in the keyword search, these instances were considered incorrect predictions.
After excluding these abnormal responses, the analysis of the remaining valid responses (those without noise) revealed the following results: 107 responses (21.57%) were deemed correct as the keywords were successfully matched, while 176 responses (35.48%) failed to match the keywords and were thus considered incorrect. An additional 42.94% of the responses were classified as "unable to locate keyword." Even after excluding the "unable to locate keyword" responses, the probability of the TinyLlama Chat Model making correct predictions that align with the factual answers was only 37.81%.
In some cases, the model generated responses that included "Garbage Text or Gibberish" unrelated to medical topics, indicating significant limitations in the TinyLlama Chat Model’s performance when handling PubMedQA questions, likely due to insufficient medical field-specific training data.
Performance Analysis of TinyLlama Chat v0.4
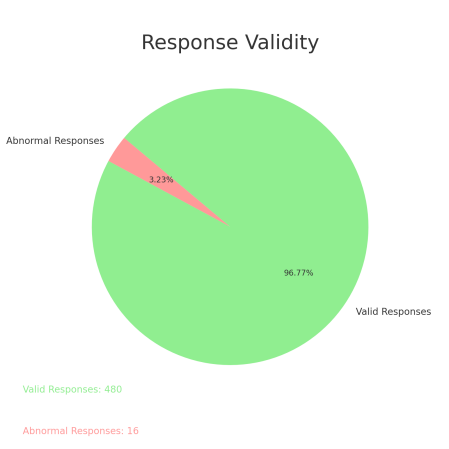
Figure 3. Pie Chart of Response Validity
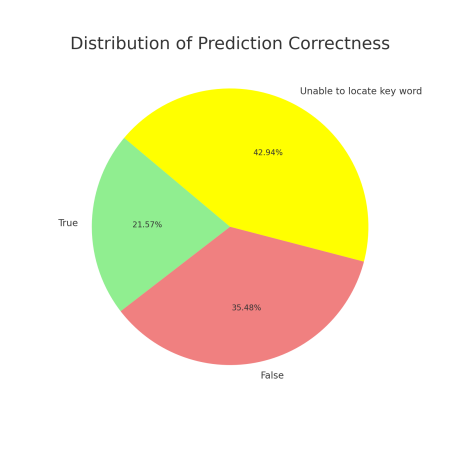
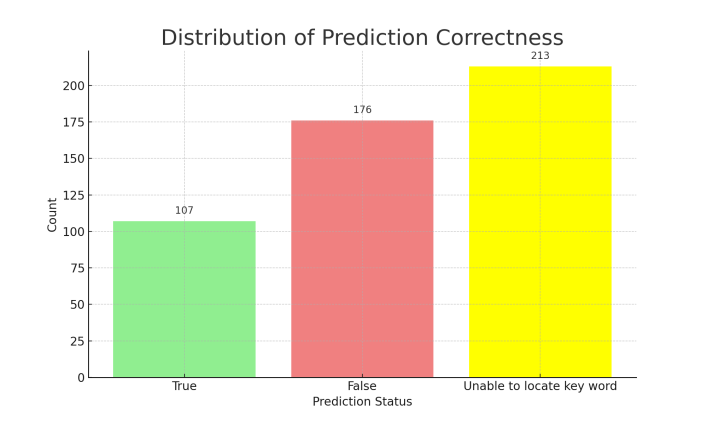
Figure 4. Pie Chart of Prediction Correctness Figure 5. Distribution of Prediction Correctness
Cura-Llama: Using the keyword detection method, the output analysis yielded the following results: 149 true responses (30.04%), 153 false responses (30.85%), and 194 responses classified as “Garbage Text or Gibberish” due to the inability to locate the keyword (39.11%). Notably, the true response rate for Cura-Llama has increased by 8.47% compared to the TinyLlama Chat v0.4.
Performance Analysis of Cura-Llama
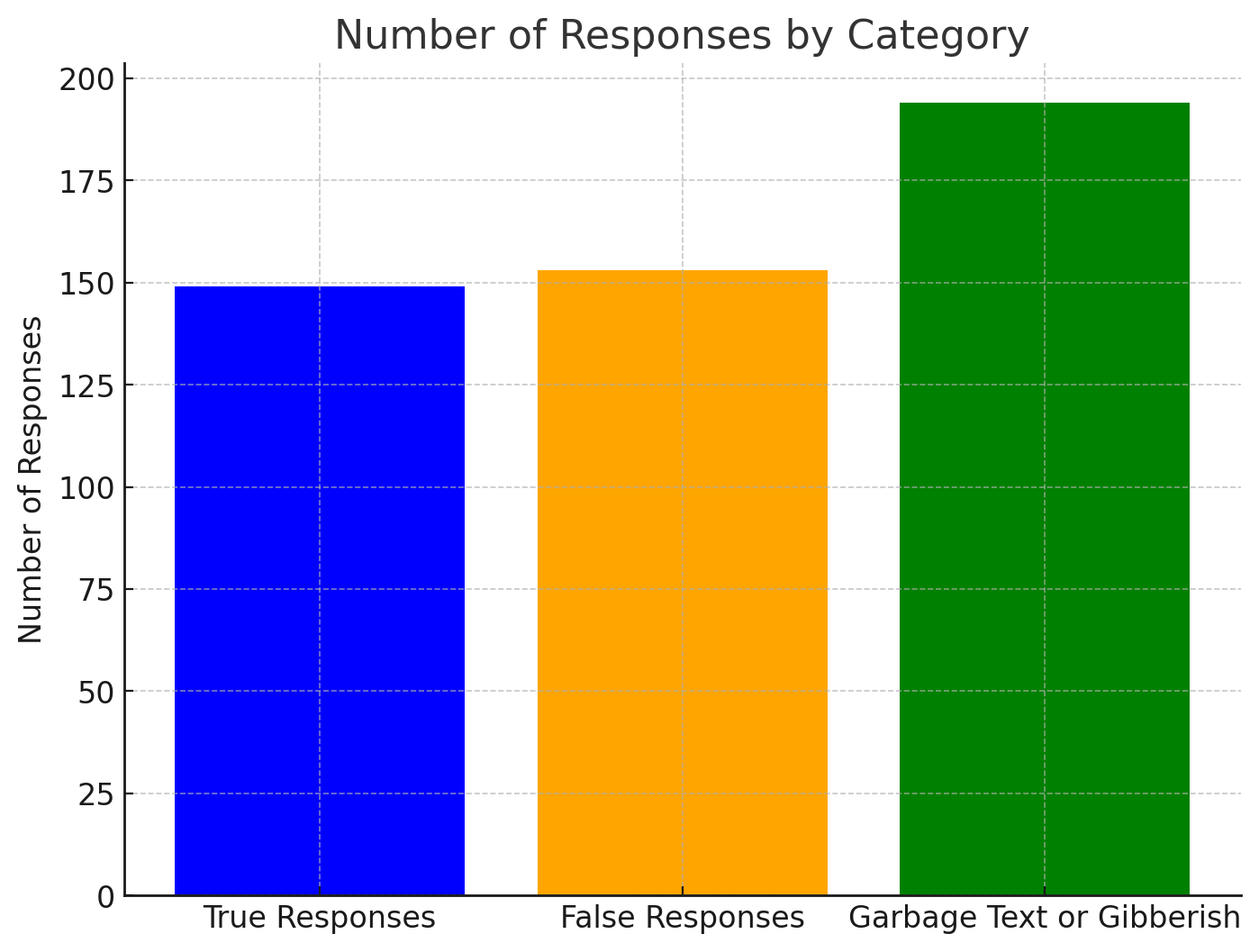
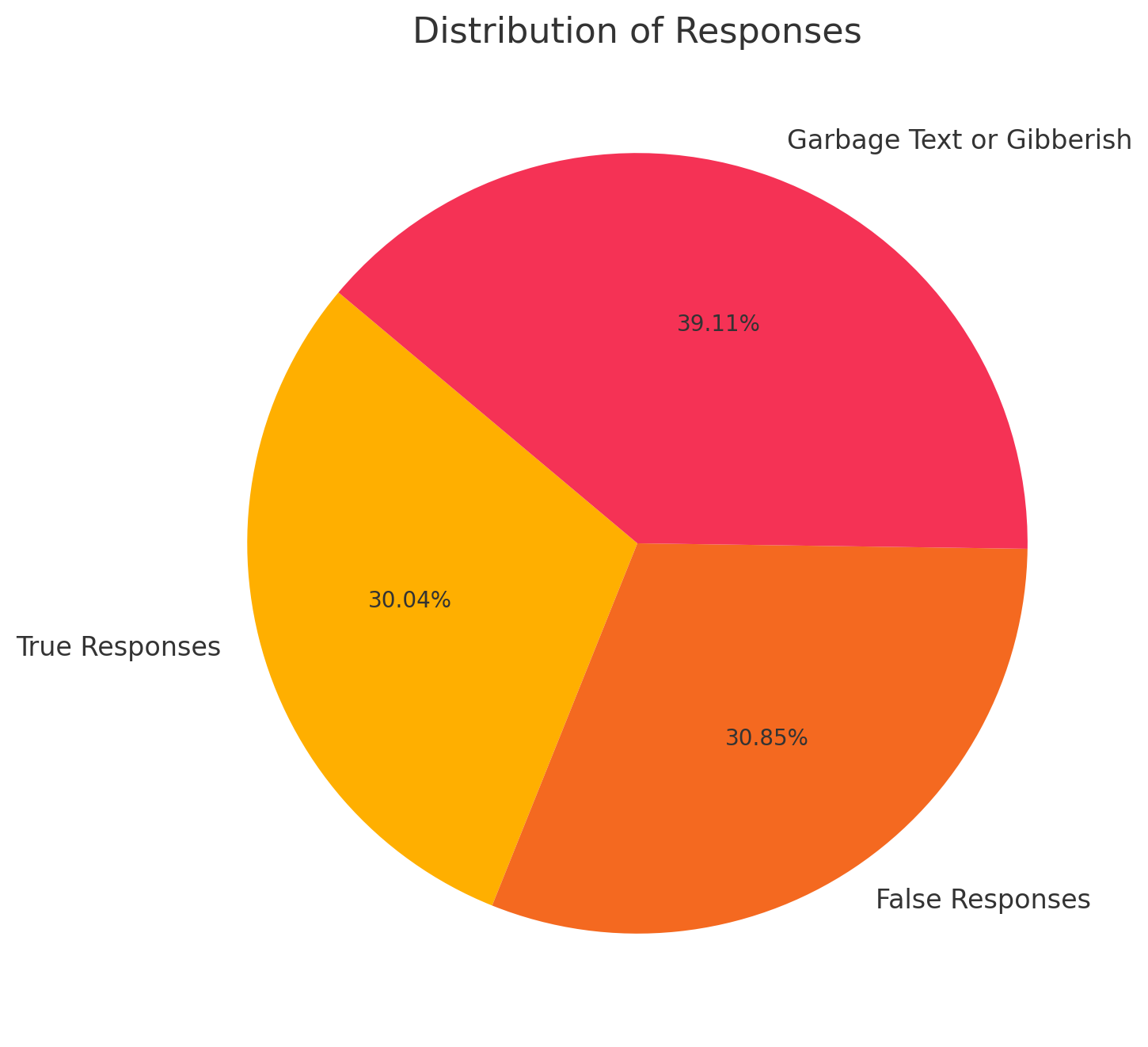
Figure 6. Distribution of Response Accuracy of Cura-Llama Figure7: Composition of Cura-LLaMA’s Responses
5. Reflection
Keyword Detection Method: The reliance on keyword detection as the primary evaluation method is a significant drawback. While straightforward, this method fails to capture the nuanced performance of the model, such as the accuracy of responses in a broader context. More sophisticated metrics, such as BERT-based evaluation or the F1 Score, could provide a more holistic assessment of the model’s performance, particularly in identifying partially correct responses or those that capture the context but lack explicit keywords.
Drawback of Omitting Other Columns in the Dataset: Another limitation is the omission of non-essential columns during the fine-tuning process. While this approach reduces computational load, it also potentially excludes valuable contextual information that could enhance the model’s understanding and response generation. Including a wider range of data features might improve the model’s ability to generate more accurate and contextually appropriate answers.
6. Conclusion
This essay demonstrates a solid effort in advancing the capabilities of open-source LLMs for medical applications. The development of Cura-LLaMA is a promising step towards making specialized AI tools more accessible, particularly in resource-constrained environments. The observed improvements in the true response rate reflect the potential benefits of fine-tuning open-source models with domain-specific data. Overall, this is a commendable attempt at addressing the unique challenges of applying LLMs in the medical field, with room for further refinement and improvement.
References
[1]. IBM. (n.d.). What are large language models? IBM. https://www.ibm.com/topics/large-language-models
[2]. Xu, Y., Su, D., & Zhang, L. (2023). Memory-efficient MoE for large language models: A dynamic routing framework. arXiv. https://arxiv.org/abs/2307.06435
[3]. Hong, F., Huang, T., Huang, X., & Shen, Z. (2023). Multimodal GPT-4 for multimodal machine reading comprehension. arXiv. https://arxiv.org/abs/2311.12351
[4]. Ma, Y., & Ye, W. (2023). Exploring temporal patterns in large language models for event detection. arXiv. https://arxiv.org/abs/2311.05232
[5]. News Medical. (2023, October 12). Large language models in medicine: Current limitations and future scope. News-Medical.net. https://www.news-medical.net/news/20231012/Large-language-models-in-medicine-Current-limitations-and-future-scope.aspx
[6]. Hugging Face. (2023, September 13). MedicalLLM leaderboard: Large language models applied to medical tasks. Hugging Face. https://huggingface.co/blog/leaderboard-medicalllm
[7]. Google Research. (n.d.). Med-PaLM: Pathways Language Model for Medicine. Google Research. https://sites.research.google/med-palm/
[8]. Pradhan, R., Liu, X., & Li, H. (2023). Prompting large language models for event coreference resolution. arXiv. https://arxiv.org/abs/2310.09089
[9]. 53AI. (2024, June 30). Large language models in AI development: The future of Qianyan technology. 53AI. https://www.53ai.com/news/qianyanjishu/2024063086075.html
[10]. Zhang, J. (2023). TinyLlama: README file. GitHub. https://github.com/jzhang38/TinyLlama/blob/main/README.md
[11]. AI Business. (2023, October). TinyLlama: The mini AI model with a trillion token punch. AI Business. https://aibusiness.com/nlp/tinyllama-the-mini-ai-model-with-a-trillion-token-punch
[12]. Hugging Face. (n.d.). PubMedQA dataset: A large-scale dataset for medical question answering. Hugging Face. https://huggingface.co/datasets/qiaojin/PubMedQA
[13]. PubMedQA. (n.d.). PubMedQA: A dataset for biomedical question answering. PubMedQA. https://pubmedqa.github.io
[14]. Zhang, J. (2023). TinyLlama: README file. GitHub. https://github.com/jzhang38/TinyLlama/blob/main/README.md
[15]. Meta AI. (n.d.). LLaMA 2: Open foundation and fine-tuned chat models. Meta AI. https://llama.meta.com/llama2/
[16]. Yadav, M., & Gupta, R. (2023). Cross-lingual transfer for dialogue systems using LLaMA 2 models. arXiv. https://arxiv.org/abs/2307.09288
[17]. Mistral AI. (2023, October). Announcing Mistral 7B: A powerful open-weight language model. Mistral. https://mistral.ai/news/announcing-mistral-7b/
[18]. Shah, A., & Huang, J. (2023). Efficient fine-tuning of large language models with adapters. arXiv. https://arxiv.org/abs/2310.06825
[19]. IBM. (n.d.). Fine-tuning models: The art of improving performance on specific tasks. IBM. https://www.ibm.com/topics/fine-tuning
[20]. LMSYS. (2023, March 30). Vicuna: An open-source chatbot for research purposes. LMSYS. https://lmsys.org/blog/2023-03-30-vicuna/
[21]. Wu, W., Tang, H., & Zhou, Y. (2023). Vicuna: Optimizing conversational agents using large language models. arXiv. https://arxiv.org/abs/2304.14454
[22]. Huang, X., Wu, J., & Liu, Z. (2024). Future challenges of multimodal AI systems. arXiv. https://arxiv.org/abs/2402.12749
[23]. Ma, J., & Zhang, L. (2024). Improving contextual understanding in LLMs through better pretraining methods. arXiv. https://arxiv.org/abs/2401.02385
[24]. Wu, T., Li, Z., & Chen, Y. (2023). Large language models for structured prediction: A case study. arXiv. https://arxiv.org/abs/2309.10818
[25]. Together AI. (2023, April). RedPajama: An open-source model for pretraining large language models. Together AI. https://www.together.ai/blog/redpajama
[26]. Hugging Face. (n.d.). Starcoder dataset: A large-scale dataset for code generation. Hugging Face. https://huggingface.co/datasets/bigcode/starcoderdata
Cite this article
Zhu,J. (2024). Cura-LLaMA: Evaluating open-source large language Model’s question answering capability on medical domain. Applied and Computational Engineering,90,52-60.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 6th International Conference on Computing and Data Science
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. IBM. (n.d.). What are large language models? IBM. https://www.ibm.com/topics/large-language-models
[2]. Xu, Y., Su, D., & Zhang, L. (2023). Memory-efficient MoE for large language models: A dynamic routing framework. arXiv. https://arxiv.org/abs/2307.06435
[3]. Hong, F., Huang, T., Huang, X., & Shen, Z. (2023). Multimodal GPT-4 for multimodal machine reading comprehension. arXiv. https://arxiv.org/abs/2311.12351
[4]. Ma, Y., & Ye, W. (2023). Exploring temporal patterns in large language models for event detection. arXiv. https://arxiv.org/abs/2311.05232
[5]. News Medical. (2023, October 12). Large language models in medicine: Current limitations and future scope. News-Medical.net. https://www.news-medical.net/news/20231012/Large-language-models-in-medicine-Current-limitations-and-future-scope.aspx
[6]. Hugging Face. (2023, September 13). MedicalLLM leaderboard: Large language models applied to medical tasks. Hugging Face. https://huggingface.co/blog/leaderboard-medicalllm
[7]. Google Research. (n.d.). Med-PaLM: Pathways Language Model for Medicine. Google Research. https://sites.research.google/med-palm/
[8]. Pradhan, R., Liu, X., & Li, H. (2023). Prompting large language models for event coreference resolution. arXiv. https://arxiv.org/abs/2310.09089
[9]. 53AI. (2024, June 30). Large language models in AI development: The future of Qianyan technology. 53AI. https://www.53ai.com/news/qianyanjishu/2024063086075.html
[10]. Zhang, J. (2023). TinyLlama: README file. GitHub. https://github.com/jzhang38/TinyLlama/blob/main/README.md
[11]. AI Business. (2023, October). TinyLlama: The mini AI model with a trillion token punch. AI Business. https://aibusiness.com/nlp/tinyllama-the-mini-ai-model-with-a-trillion-token-punch
[12]. Hugging Face. (n.d.). PubMedQA dataset: A large-scale dataset for medical question answering. Hugging Face. https://huggingface.co/datasets/qiaojin/PubMedQA
[13]. PubMedQA. (n.d.). PubMedQA: A dataset for biomedical question answering. PubMedQA. https://pubmedqa.github.io
[14]. Zhang, J. (2023). TinyLlama: README file. GitHub. https://github.com/jzhang38/TinyLlama/blob/main/README.md
[15]. Meta AI. (n.d.). LLaMA 2: Open foundation and fine-tuned chat models. Meta AI. https://llama.meta.com/llama2/
[16]. Yadav, M., & Gupta, R. (2023). Cross-lingual transfer for dialogue systems using LLaMA 2 models. arXiv. https://arxiv.org/abs/2307.09288
[17]. Mistral AI. (2023, October). Announcing Mistral 7B: A powerful open-weight language model. Mistral. https://mistral.ai/news/announcing-mistral-7b/
[18]. Shah, A., & Huang, J. (2023). Efficient fine-tuning of large language models with adapters. arXiv. https://arxiv.org/abs/2310.06825
[19]. IBM. (n.d.). Fine-tuning models: The art of improving performance on specific tasks. IBM. https://www.ibm.com/topics/fine-tuning
[20]. LMSYS. (2023, March 30). Vicuna: An open-source chatbot for research purposes. LMSYS. https://lmsys.org/blog/2023-03-30-vicuna/
[21]. Wu, W., Tang, H., & Zhou, Y. (2023). Vicuna: Optimizing conversational agents using large language models. arXiv. https://arxiv.org/abs/2304.14454
[22]. Huang, X., Wu, J., & Liu, Z. (2024). Future challenges of multimodal AI systems. arXiv. https://arxiv.org/abs/2402.12749
[23]. Ma, J., & Zhang, L. (2024). Improving contextual understanding in LLMs through better pretraining methods. arXiv. https://arxiv.org/abs/2401.02385
[24]. Wu, T., Li, Z., & Chen, Y. (2023). Large language models for structured prediction: A case study. arXiv. https://arxiv.org/abs/2309.10818
[25]. Together AI. (2023, April). RedPajama: An open-source model for pretraining large language models. Together AI. https://www.together.ai/blog/redpajama
[26]. Hugging Face. (n.d.). Starcoder dataset: A large-scale dataset for code generation. Hugging Face. https://huggingface.co/datasets/bigcode/starcoderdata









