1. Introduction
Brain tumors, as one of the most common life-threatening neurological disorder of the nervous system, have become a significant global public health concern [1]. According to statistics from the World Health Organization, the annual incidence of brain tumors continues to rise, particularly among the middle-aged and elderly populations, where the increase is especially pronounced. Although therapeutic approaches for brain tumors have steadily advanced, many patients are often diagnosed at a late stage due to the insidious nature of early symptoms. This results in a shortened therapeutic window and poor prognosis. Therefore, early and accurate detection of brain tumors is of critical importance for improving patient survival rates, enhancing treatment outcomes, and prolonging life expectancy.
Magnetic Resonance Imaging (MRI), as a non-invasive imaging technique, is widely used in the diagnosis of brain diseases due to its ability to provide superior soft-tissue contrast of brain tissues. MRI not only accurately depicts the location, morphology, and spatial relationship of brain tumors with surrounding tissues but also avoids exposure to ionizing radiation, offering substantial safety benefits for patients. As such, MRI imaging plays an irreplaceable role in brain tumor detection and is currently one of the most commonly used diagnostic tools in clinical practice.
Although MRI technology provides reliable imaging data for brain tumor detection, traditional diagnostic methods based on subjective interpretation and manual annotation by radiologists still present notable limitations. Radiologists typically identify and classify tumors based on visual appearance, morphological features, and lesion distribution. This process relies heavily on the clinician’s expertise and experience. However, the imaging characteristics of brain tumors often exhibit high heterogeneity—tumor shape, size, location, and interaction with adjacent brain structures can vary significantly, making interpretation more complex. Moreover, tumor boundaries are sometimes ambiguous, and low-contrast lesions may be difficult to distinguish from normal brain tissue, placing high demands on diagnostic accuracy.
Manual interpretation and annotation are not only experience-dependent but also susceptible to subjectivity, which can lead to variability in diagnostic results among different clinicians, ultimately affecting the consistency and accuracy of diagnosis. These limitations are particularly evident in cases involving diverse tumor types, small lesions, or tumors located in anatomically challenging regions of the brain.
Although traditional machine learning methods have been widely applied in medical image analysis, employing manually designed feature extraction and classification algorithms for preliminary tumor identification and classification, they still face many challenges. Traditional machine learning approaches rely on hand-crafted features, which often fail to comprehensively capture the complex characteristics of brain tumors in MRI images. This is especially problematic when dealing with diverse tumor types, low-contrast lesions, and indistinct boundaries, all of which significantly impact classification performance. Furthermore, these methods often require extensive domain expertise and incur high computational costs, making them inefficient for processing large-scale imaging data and incapable of delivering high-precision results within a short time frame. These constraints make traditional imaging-based diagnostic approaches insufficient to meet the clinical demand for high-accuracy and high-efficiency brain tumor detection.
Over recent years, the rapid evolution of deep learning—particularly Convolutional Neural Networks (CNN)—has brought revolutionary progress to the field of medical image analysis [2]. As a powerful image processing tool, CNNs can automatically learn effective features from large-scale MRI datasets and extract deep hierarchical information through multi-layer network architectures. Unlike traditional methods, CNNs do not require manually designed features but instead autonomously identify key regions and details in medical images, thereby greatly improving the accuracy and robustness of image recognition. CNNs have demonstrated strong adaptability and superior performance in handling high-dimensional, complex, and heterogeneous medical images. By training on large volumes of imaging data, CNNs can identify underlying patterns and relationships, enabling accurate brain tumor detection and significantly enhancing classification and segmentation performance.
This study aims to explore brain tumor detection methods based on artificial intelligence, specifically focusing on the application of CNNs in MRI image analysis. Through deep learning analysis of MRI data and the development of advanced CNN models, this paper investigates approaches to improve the accuracy and efficiency of brain tumor detection, providing a more precise and effective auxiliary tool for clinical diagnosis.
2. Previous works
Traditional image classification models typically rely on manual feature extraction and employ comparatively basic algorithms for classification tasks. Although such models can perform adequately under certain conditions, they often encounter considerable difficulties when dealing with complex data. Common traditional image classification methods include K-Nearest Neighbors (KNN), Support Vector Machines (SVM), Decision Trees, and Random Forests. In brain tumor detection, these models usually require manual preprocessing and feature extraction before the image data can be fed into the classification algorithm.
KNN is an instance-based learning method that classifies samples by calculating the distance—usually Euclidean or Manhattan distance—between the test sample and the training samples [3]. Before being input into the KNN algorithm, images must first undergo feature extraction using techniques such as edge detection or color histograms to transform the raw image into a feature vector. While KNN is simple and intuitive, it is susceptible to the "curse of dimensionality" in high-dimensional data, which can lead to decreased classification performance.
SVM performs classification by constructing a hyperplane that separates different categories. Input images must first undergo feature extraction, commonly through techniques like Principal Component Analysis (PCA) or local feature descriptors such as Scale-Invariant Feature Transform (SIFT) or Histogram of Oriented Gradients (HOG). SVM is well-suited for small sample datasets, but its performance is highly sensitive to parameter selection, such as the choice of kernel function and penalty parameters. Moreover, the computational cost becomes significant when handling large-scale datasets.
Decision Trees perform classification through an iterative partitioning process that builds a tree-like hierarchy, with each node corresponding to specific features and its branches indicating potential value ranges. Image data must also be transformed through feature extraction before being used in a decision tree model. Decision Trees offer good interpretability and can clearly illustrate the decision-making process. However, they are prone to overfitting, which can degrade performance on unseen data.
Random Forests enhance classification accuracy by aggregating multiple decision trees. Each tree is trained using a random subset of features, which effectively mitigates overfitting. However, similar to decision trees, Random Forests require feature extraction prior to model input, typically using the same methods. Although Random Forests generally achieve high classification performance, they come with increased model complexity, leading to longer training and prediction times.
Traditional classification models heavily rely on manual feature extraction when processing image data. This process is time-consuming and requires domain expertise to identify appropriate features, which limits the models' performance when confronted with complex image characteristics. For instance, in brain tumor MRI images, variations in tumor shape, texture, and surrounding tissue can significantly influence classification accuracy. Traditional models often struggle to capture such complex spatial relationships and subtle variations, leading to reduced classification performance. Additionally, these models face challenges in computational efficiency and accuracy when processing large-scale datasets. As the volume of MRI images increases, both training and inference times for traditional algorithms grow substantially, potentially resulting in wasted computational resources and performance bottlenecks. Moreover, parameter tuning in traditional models often depends on prior knowledge and lacks adaptability, further limiting their practical applicability.
In contrast, deep learning, particularly CNNs, can automatically extract multi-level features directly from raw images, overcoming the limitations of traditional methods. Through the stacking of multiple convolutional and pooling layers, CNNs can effectively capture spatial structures and local patterns within images, thereby achieving more accurate classification. In the context of brain tumor detection from MRI images, CNNs not only recognize tumor boundaries and morphological variations but also learn more complex feature representations, enhancing the robustness of classification performance.
3. Dataset and preprocessing
A brain tumor refers to a mass or collection of abnormal cells in the brain, which can be either cancerous (malignant) or non-cancerous (benign). In this study, a merged dataset from multiple sources was used for comprehensive analysis [4], [5], [6], [7]. The dataset consists of 7,023 human brain MRI images categorized into four classes: glioma, meningioma, no tumor, and pituitary tumor (Figure 1).
Figure 1: Samples from the dataset used in this study
The training set used in this study comprises MRI images from the four primary categories of brain tumors. First, the glioma class includes 1,321 MRI images of glioma tumors. As a malignant type of brain tumor with diverse morphologies and characteristics, these images are crucial for enabling the model to learn the specific features of gliomas. Second, the meningioma class contains 1,339 MRI images. Meningiomas are typically benign tumors whose location and imaging features differ from those of gliomas, making them significant for accurate classification and detection. Additionally, the no tumor class includes 1,595 MRI images of healthy brain tissues, serving as a critical reference for the model to improve classification accuracy and reduce misdiagnosis. Finally, the pituitary tumor class comprises 1,457 MRI images. Pituitary tumors, located in the pituitary gland, exhibit unique imaging characteristics, and accurate classification of these images is essential for clinical diagnosis. In the test set, the number of images for each category is as follows: 300 (glioma), 306 (meningioma), 405 (no tumor), and 300 (pituitary).
To address the scarcity of medical imaging data and enhance the generalization ability of the model, this study employed data augmentation techniques. A composite augmentation strategy was designed for the training set: first, the original MRI images were subjected to random horizontal flip and vertical flip to simulate imaging variations from different scanning orientations. Subsequently, random rotations within ±10° were applied to improve the model’s robustness to directional variations of lesions. All images were uniformly resized to 224×224 pixels, followed by tensor transformation and normalization. The normalization parameters followed the ImageNet standard (mean: [0.485, 0.456, 0.406]; standard deviation: [0.229, 0.224, 0.225]).
For the validation and test sets, a deterministic preprocessing pipeline was applied, which included only resizing, tensor transformation, and normalization, in order to eliminate the randomness introduced by data augmentation and ensure fair model performance evaluation. The dataset was split into training and test sets in a 7:3 ratio. Within the training set, 10% of the data was further set aside as a validation set for hyperparameter tuning.
4. Model
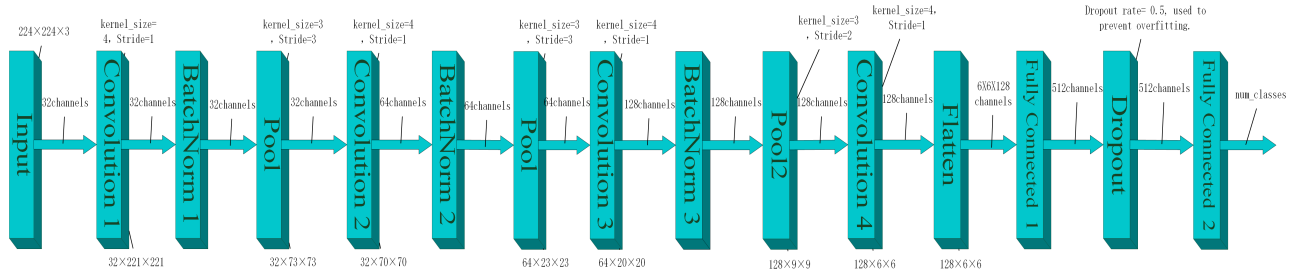
Figure 2: Model structure
The CNN architecture designed in this study consists of three integrated modules: the feature extraction module, the spatial down-sampling module, and the classification decision module (as illustrated in Figure 2).
The feature extraction module comprises four progressive convolutional layers responsible for extracting multi-scale lesion features from the input MRI images. The first convolutional layer employs kernels of size 4×4 with 32 output channels. The number of channels increases in subsequent layers following a 64-128-128 progression. After each convolution operation, Batch Normalization and ReLU non-linear activation are applied sequentially to accelerate model convergence and enhance the expressiveness of complex texture features. All convolutional layers adopt a stride of 1 and no padding (padding=0), maintaining sensitivity to local features while minimizing the number of parameters. The spatial down-sampling module is composed of multi-level pooling layers embedded within the feature extraction process to achieve progressive spatial compression. The outputs of the first three convolutional layers are followed by 3×3 Max Pooling layers with a stride of 3, preserving significant feature responses while suppressing noise. The fourth layer incorporates an improved pooling operation (kernel=3×3, stride=2), reducing the feature map resolution to 6×6 while mitigating excessive loss of small lesion information. The classification decision module is responsible for final classification. The 128-channel feature maps, after spatial compression, are flattened and passed into a fully connected layer, which maps the features to a 512-dimensional latent space. This layer enables high-level feature interactions to establish correlations between lesion representations and class labels. To prevent overfitting, a Dropout layer with a dropout rate of 50% is applied after the hidden layer, randomly deactivating some neurons to enhance generalization capability. The final output layer utilizes a Softmax function to produce a four-dimensional probability vector corresponding to the healthy group and the three tumor subtypes, completing the end-to-end classification task.
During the training phase, the Adam optimizer was adopted with an initial learning rate set to 0.0002, and cross-entropy loss was selected as the loss function. In each training batch, forward propagation is followed by gradient zeroing and backpropagation, where model parameters are dynamically updated to minimize prediction error. To monitor the training process, cumulative loss and classification accuracy are calculated in real time after every 150 batch updates. Upon completion of the full training cycle, average training loss and accuracy are recorded as indicators of model convergence. Additionally, an Early Stopping mechanism was introduced to enhance generalization performance: if the validation loss fails to decrease for five consecutive epochs, training is automatically terminated to avoid overfitting.
Through this design, the model achieves a balanced trade-off between feature representation capacity and generalization ability, effectively addressing the challenges of overfitting in complex medical image classification tasks.
5. Results
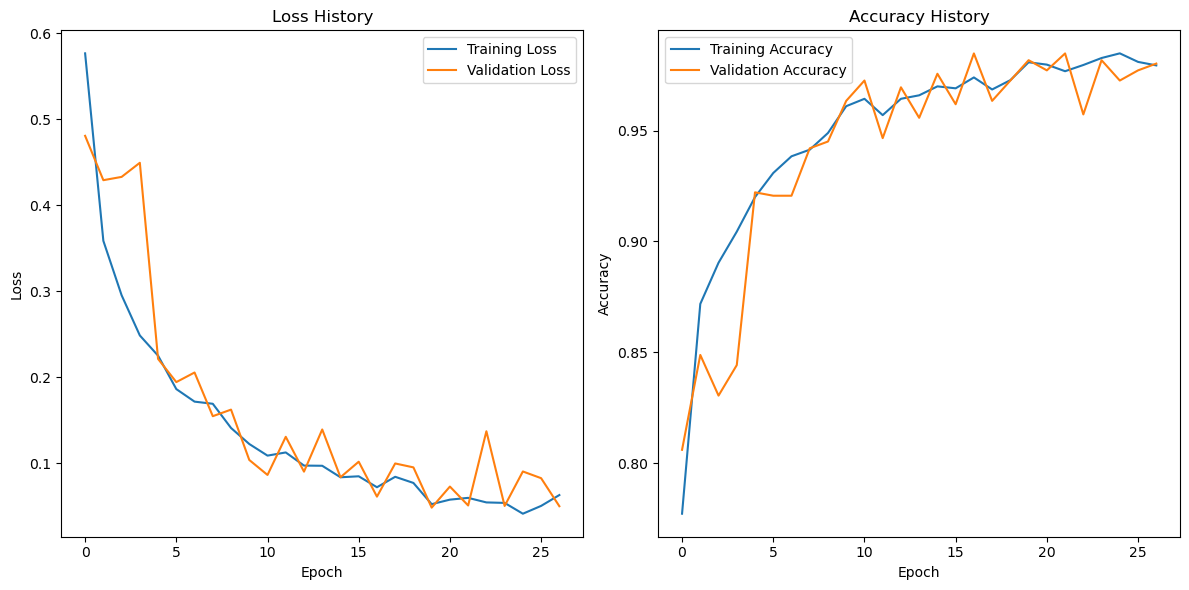
Figure 3: Model loss and accuracy curve
During the model training process, the loss function exhibited an ideal convergence trend. From the training loss curve, it can be observed that the loss value decreased rapidly as the number of training epochs increased. In the initial epochs, the training loss dropped significantly from approximately 0.55 to around 0.2, and then gradually stabilized throughout the later stages of training, eventually reaching a low and steady value of approximately 0.07. The validation loss followed a similar downward trend. Although slight fluctuations occurred in certain epochs, the overall pattern closely mirrored that of the training loss, ultimately stabilizing at around 0.08. This indicates that the model achieved good fitting performance on both the training and validation sets, without exhibiting significant signs of overfitting.
The trends in training and validation accuracy further confirm the effectiveness of the model. The training accuracy started at approximately 0.80 and steadily increased as training progressed, surpassing 0.95 after around 10 epochs and eventually stabilizing at approximately 0.96. The validation accuracy also demonstrated strong performance. Despite some minor fluctuations during training, it exhibited a general upward trend and ultimately stabilized at approximately 0.95, closely aligning with the training accuracy. These results further demonstrate the model’s strong generalization ability and performance stability.
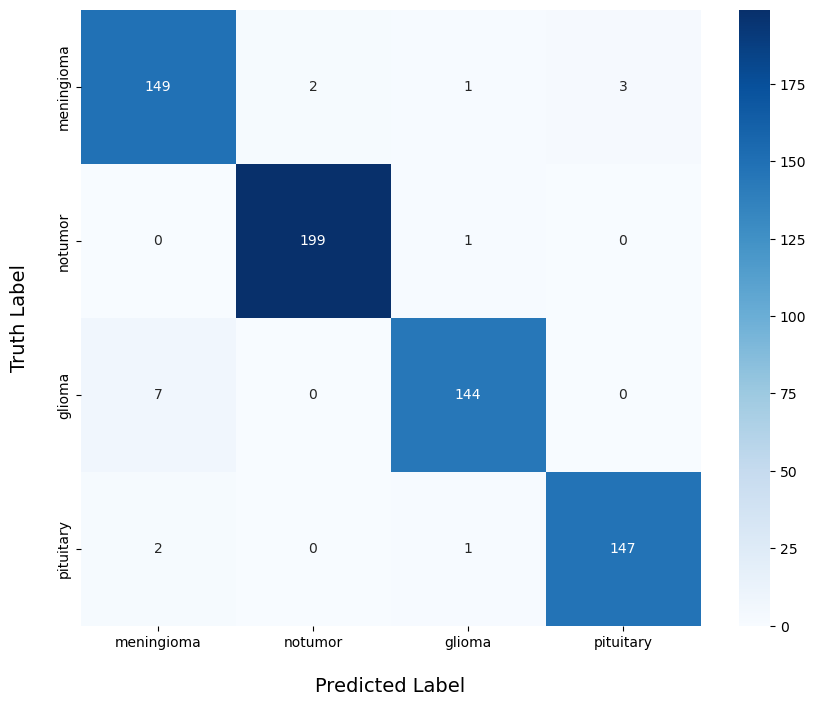
Figure 4: Prediction results
Regarding the model’s actual classification performance, the confusion matrix provides an intuitive visualization of its effectiveness across different categories. For the “notumor” class, the model demonstrated exceptionally high accuracy, with only 1 out of 199 samples misclassified, resulting in a correct classification rate of 99.5%. This indicates that the model can reliably distinguish non-tumor cases with a high degree of confidence. In the “meningioma” category, the model also performed well, correctly classifying 149 out of 155 samples, yielding an accuracy of 96.1%. Only a small number of samples were misclassified into other categories. For the “pituitary” class, 147 out of 150 samples were correctly identified, achieving an accuracy rate of 98%, which demonstrates the model’s strong discriminative ability for this tumor type.
However, the model showed slightly weaker performance in the “glioma” category. Among 151 samples, 144 were correctly classified, with an accuracy rate of 95.4%. Although this is still a high accuracy rate, it is relatively lower compared to the other classes, with a higher number of misclassifications. This may be attributed to the greater complexity of glioma tumor features or the higher variability within the glioma samples. These findings suggest that further optimization and refinement in handling this specific category could help improve the overall classification performance in future research.
6. Discussion and conclusion
This study proposed an artificial intelligence-based brain tumor detection method using MRI images, achieving automatic classification and recognition of brain tumors through a deep learning model. Experimental results demonstrated that the model performed well during both training and validation phases. The training and validation losses showed a steadily decreasing trend, eventually converging to approximately 0.07 and 0.08, respectively. Meanwhile, the training and validation accuracies stabilized at around 0.96 and 0.95, indicating the model’s high precision in data fitting and its strong generalization capability on unseen validation data.
Further evaluation through confusion matrix analysis revealed that the model achieved high classification accuracy across various categories. Specifically, the correct classification rates for the “notumor,” “meningioma,” and “pituitary” classes reached 99.5%, 96.1%, and 98%, respectively. Although the performance for the “glioma” class was slightly lower, the model still achieved an accuracy of 95.4%, indicating overall excellent classification performance.
However, the model still has several limitations. Firstly, the model architecture is relatively simple, which may hinder its ability to capture all critical features when dealing with more complex or highly heterogeneous tumor imaging data, thus affecting classification accuracy. Secondly, although the model exhibited good robustness on the current dataset, its generalization ability needs further validation and enhancement—especially when applied to MRI images from different medical institutions, scanning equipment, or with varying imaging parameters, where adaptability issues may arise. From a computational efficiency perspective, while the current training and inference times are within acceptable limits, there is still room for improvement in processing large-scale datasets or performing real-time detection tasks.
In the future, several strategies can be employed to optimize deep learning-based brain tumor MRI image classification methods. Firstly, more advanced network architectures such as Transformers can be introduced to capture more global image features [8], or transfer learning can be leveraged to utilize large-scale pretrained models [9], thereby improving generalization and reducing data requirements. Secondly, incorporating multimodal imaging data (e.g., MRI, PET, CT) can provide complementary information from different imaging modalities, enhancing diagnostic accuracy. Efficient training algorithms such as distributed training and self-supervised learning can also be explored to reduce reliance on large amounts of labeled data, thereby lowering computational costs and improving adaptability. Data augmentation techniques [10], such as generating synthetic images with GANs [11], can expand the training dataset and improve recognition of rare tumor types.
Further clinical validation studies—particularly large-scale, multi-center trials—are essential to ensure the model’s stability and applicability across diverse populations, imaging devices, and acquisition parameters. Enhancing the interpretability of AI diagnostics through techniques such as attention-based visualization can help clinicians better understand the model’s decision-making process and build trust in AI systems. In addition, integrating pathological, genomic, and other interdisciplinary data can facilitate precision medicine and support personalized treatment strategies.
Optimizing the integration of AI diagnostic systems into existing hospital information infrastructures (e.g., HIS, PACS) can improve the convenience and efficiency of clinical applications. Attention should also be given to model scalability to accommodate emerging tumor subtypes and evolving imaging technologies. Finally, fostering interdisciplinary collaboration among computer scientists, radiologists, pathologists, and clinicians is crucial to accelerating the clinical translation of AI diagnostic technologies and ultimately improving the quality of care for brain tumor patients.
References
[1]. S. Bauer, R. Wiest, L.-P. Nolte, and M. Reyes, “A survey of MRI-based medical image analysis for brain tumor studies,” Phys. Med. Biol., vol. 58, no. 13, p. R97, Jun. 2013, doi: 10.1088/0031-9155/58/13/R97.
[2]. Z. Li, F. Liu, W. Yang, S. Peng, and J. Zhou, “A survey of convolutional neural networks: Analysis, applications, and prospects,” IEEE Transactions on Neural Networks and Learning Systems, vol. 33, no. 12, pp. 6999–7019, Dec. 2022, doi: 10.1109/TNNLS.2021.3084827.
[3]. V. Wasule and P. Sonar, “Classification of brain MRI using SVM and KNN classifier,” in 2017 Third International Conference on Sensing, Signal Processing and Security (ICSSS), May 2017, pp. 218–223. doi: 10.1109/SSPS.2017.8071594.
[4]. J. Cheng et al., “Enhanced performance of brain tumor classification via tumor region augmentation and partition,” PLOS ONE, vol. 10, no. 10, p. e0140381, Oct. 2015, doi: 10.1371/journal.pone.0140381.
[5]. J. Cheng et al., “Retrieval of brain tumors by adaptive spatial pooling and fisher vector representation,” PLOS ONE, vol. 11, no. 6, p. e0157112, Jun. 2016, doi: 10.1371/journal.pone.0157112.
[6]. Sartaj Bhuvaji, Ankita Kadam, Prajakta Bhumkar, Sameer Dedge, and Swati Kanchan, “Brain tumor classification (MRI).” Kaggle. doi: 10.34740/KAGGLE/DSV/1183165.
[7]. Msoud Nickparvar, “Brain tumor MRI dataset.” Kaggle. doi: 10.34740/KAGGLE/DSV/2645886.
[8]. P. Wang, Q. Yang, Z. He, and Y. Yuan, “Vision transformers in multi-modal brain tumor MRI segmentation: A review,” Meta-Radiology, vol. 1, no. 1, p. 100004, Jun. 2023, doi: 10.1016/j.metrad.2023.100004.
[9]. C. Srinivas et al., “Deep transfer learning approaches in performance analysis of brain tumor classification using MRI images,” Journal of Healthcare Engineering, vol. 2022, no. 1, p. 3264367, 2022, doi: 10.1155/2022/3264367.
[10]. J. Nalepa, M. Marcinkiewicz, and M. Kawulok, “Data augmentation for brain-tumor segmentation: A review,” Front. Comput. Neurosci., vol. 13, Dec. 2019, doi: 10.3389/fncom.2019.00083.
[11]. C. Han et al., “Combining noise-to-image and image-to-image GANs: Brain MR image augmentation for tumor detection,” IEEE Access, vol. 7, pp. 156966–156977, 2019, doi: 10.1109/ACCESS.2019.2947606.
Cite this article
Li,Q. (2025). Brain Tumor Detection Based on MRI Images and Artificial Intelligence. Applied and Computational Engineering,139,67-75.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 7th International Conference on Computing and Data Science
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. S. Bauer, R. Wiest, L.-P. Nolte, and M. Reyes, “A survey of MRI-based medical image analysis for brain tumor studies,” Phys. Med. Biol., vol. 58, no. 13, p. R97, Jun. 2013, doi: 10.1088/0031-9155/58/13/R97.
[2]. Z. Li, F. Liu, W. Yang, S. Peng, and J. Zhou, “A survey of convolutional neural networks: Analysis, applications, and prospects,” IEEE Transactions on Neural Networks and Learning Systems, vol. 33, no. 12, pp. 6999–7019, Dec. 2022, doi: 10.1109/TNNLS.2021.3084827.
[3]. V. Wasule and P. Sonar, “Classification of brain MRI using SVM and KNN classifier,” in 2017 Third International Conference on Sensing, Signal Processing and Security (ICSSS), May 2017, pp. 218–223. doi: 10.1109/SSPS.2017.8071594.
[4]. J. Cheng et al., “Enhanced performance of brain tumor classification via tumor region augmentation and partition,” PLOS ONE, vol. 10, no. 10, p. e0140381, Oct. 2015, doi: 10.1371/journal.pone.0140381.
[5]. J. Cheng et al., “Retrieval of brain tumors by adaptive spatial pooling and fisher vector representation,” PLOS ONE, vol. 11, no. 6, p. e0157112, Jun. 2016, doi: 10.1371/journal.pone.0157112.
[6]. Sartaj Bhuvaji, Ankita Kadam, Prajakta Bhumkar, Sameer Dedge, and Swati Kanchan, “Brain tumor classification (MRI).” Kaggle. doi: 10.34740/KAGGLE/DSV/1183165.
[7]. Msoud Nickparvar, “Brain tumor MRI dataset.” Kaggle. doi: 10.34740/KAGGLE/DSV/2645886.
[8]. P. Wang, Q. Yang, Z. He, and Y. Yuan, “Vision transformers in multi-modal brain tumor MRI segmentation: A review,” Meta-Radiology, vol. 1, no. 1, p. 100004, Jun. 2023, doi: 10.1016/j.metrad.2023.100004.
[9]. C. Srinivas et al., “Deep transfer learning approaches in performance analysis of brain tumor classification using MRI images,” Journal of Healthcare Engineering, vol. 2022, no. 1, p. 3264367, 2022, doi: 10.1155/2022/3264367.
[10]. J. Nalepa, M. Marcinkiewicz, and M. Kawulok, “Data augmentation for brain-tumor segmentation: A review,” Front. Comput. Neurosci., vol. 13, Dec. 2019, doi: 10.3389/fncom.2019.00083.
[11]. C. Han et al., “Combining noise-to-image and image-to-image GANs: Brain MR image augmentation for tumor detection,” IEEE Access, vol. 7, pp. 156966–156977, 2019, doi: 10.1109/ACCESS.2019.2947606.









