1. Introduction
The early development of drosophila contains a series of process that relies on the expression of genes, which can be regulated by multiple transcription factors. The drosophila embryo experiences an essential process maternal-to-zygotic transformation (MZT), this requires zygotic genome activation (ZGA) and degradation of maternal products [1].
It was reported that Zld protein is one of those transcription factors that promotes zygotic genome activation [2]. It binds on a specific region CAGTAG to promote transcription. The binding site can be found by Chromatin Immuno-Precipitation, which links protein to DNA and detect whether and where a protein binds to Chromatin [3].
In this study, the editing was done by mutating the Zld binding site of CG15382. Target gene were expected to be silenced without the expression of the activator. In situ hybridization were expected to be done to show the expression of CG15382 in mutants and normal embryos. Finally, the function of CG15382 was studied by analyzing the differences between mutant Drosophila embryos and normal embryos. The expression of CG15382 is predicted to be decreased in the Zld absent mutant.
2. MATERIALS
2.1. Flybase
Gene data (including its length, sequence, etc.) were collected from Flybase website to create the guide RNA. Sequences were copied to other software to get PAM sequence and CAGGTAG sequence.
2.2. ChopChop & SnapGene:
Two software were used to design the guide RNA, which is ChopChop and SnapGene.
ChopChop was used to find the PAM sequence, after inputting about 50 base pairs around a target sequence onto the website, it will automatically search for the part containing the PAM sequence. Eventually 6 sequences were found, the PAM sequence that is closest to the target sequence and does not contain the CAGTAG sequence is selected as the guide RNA.
SnapGene is a software that helps to create the guide RNA. The sequence of CG15382 were imported into the software, then the guide RNA sequence was designed in the SnapGene.
2.3. Integrated Genome Browesr (IGB)
IGB is a software that used to analyze the results of ChIP. After importing the experimental data into the software, advanced search were used to locate the CG15382 gene. By combining the binding status of proteins with the position of gene sequences for analysis, the binding sites of Zld can be identified.
3. Methods
3.1. Chromatin Immuno-Precipitation (ChIP)
To determine the binding site of Zld, ChIP was used to show the binding of Zld protein to DNA sequence. Experiments were done by Y.M., the data are from Rushlow unpublished results.
The principle of ChIP is to determine which specific DNA sequences are associated with specific proteins to reveal their role in regulating gene expression. Cells are processed under physiological conditions to form covalent cross-linking between proteins and DNA. Then the cells were lysed, and its DNA was cut through ultrasound or enzyme digestion, producing small fragments of DNA containing target protein binding. After that, specific antibodies were added to DNA to bind the target protein. Other proteins that can bind to the antibody were also added to precipitate the Chromatin fragment containing the target protein. After reverse crosslinking treatment, protein and DNA are separated, and then DNA is extracted and purified. Finally, analyze the obtained DNA [4].
3.2. CRISPR-Cas9:
To mutate the binding sequence, CRISPR-Cas9 editing was designed.
Since the DNA sequence has two binding sequences, two guide RNA were designed to replace the original sequence. A sequence with approximately 50 bp around the CAGGTAG sequence was selected and the chopshop website was used to locate the sequence containing PAM. The cutting site should not contain the CAGGTAG sequence. The guide RNA was designed as a substitute sequence with 20 base pairs and two homologous sequences with 500 base pairs on each side of the target sequence. The substitute sequence is a 20 base pairs sequence upstream of the PAM, but the target sequence was replaced, the CAGGTAG sequence were designed to be replaced by CTCATAG.
The gene editing is expected to be done by CRSIPR-Cas9. Two plasmids will be injected into drosophila embryo, one expresses the DNA cleaving protein Cas9, and the other expresses guide RNA. The guide RNA sequence will first bind the target sequence, then the Cas9 protein will cleave the DNA sequence at the position of three base pairs upstream of the PAM. The editing is made by the principle of homology directed repair (HDR), which is known as a process used by cells to accurately repair breaks that occur on both strands of DNA. The repair occurs in the present of a template RNA, and the recombination occurs between the homologous regions. After the injected plasmid expresses guide RNA and Cas9 protein, the homologous sequence in guide RNA will first control the binding of guide RNA to the target sequence then the Cas9 enzyme introduce a double-strand break at a specific site in the genome, the target sequence will be replaced by the guide RNA [5].
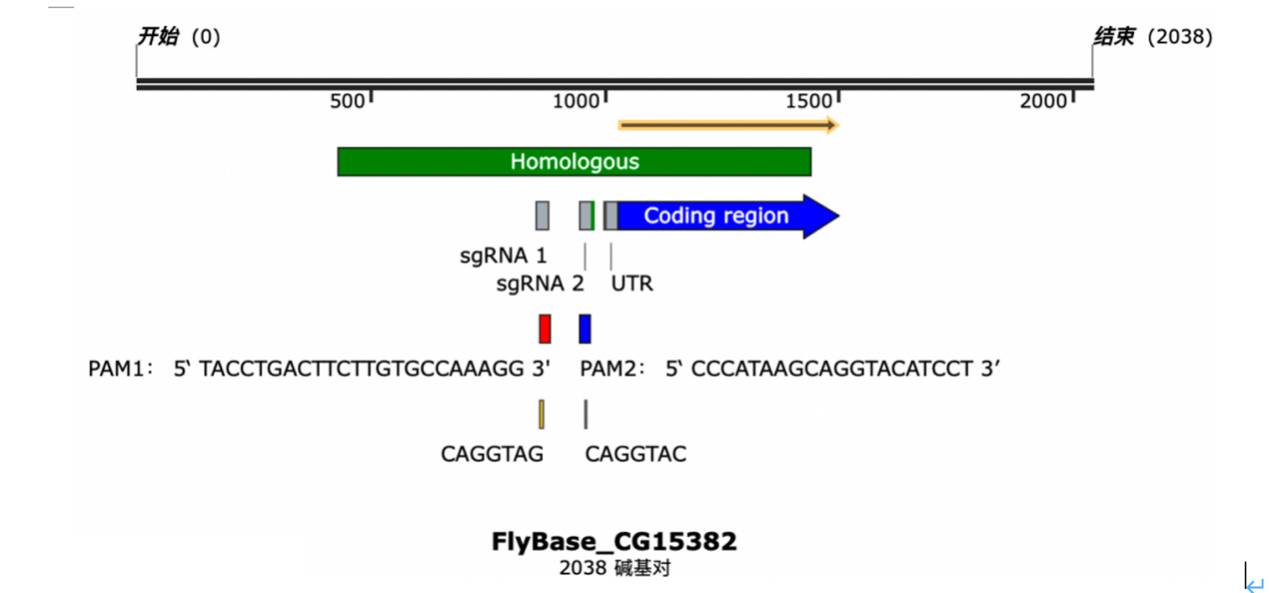
Figure 1. The sketch of the sgRNA
The two sgRNA shares the same homologous sequence, but the PAM sequence are individually shown. UTR are the abbreviation of untranslated region. The coding region are also marked above.

Figure 2. The PAM sequence
The PAM sequence of both two sgRNA are shown. The binding sequence of Zld and its replacing sequence are also shown in yellow.
The two sgRNA were designed as above.
3.3. In situ hybridization:
In situ hybridization is performed by Y.M.. The purpose of in situ hybridization is to obtain stained Drosophila embryos.
In situ hybridization (ISH) is an experimental method for locating specific nucleic acid sequences in cell or tissue sections. The main steps include preparing a probe (usually a labeled piece of DNA or RNA that can specifically pair with the target sequence), hybridization (the pairing process between the probe and the target sequence), washing [6].
Overall, to study the spatial and temporal distribution of genes in cells and tissues, in situ hybridization was done to the Drosophila embryos.
4. Result
4.1. CG15382 contains two Zld binding sites
The binding of polymerase to the DNA sequence boost at nucleus cycle 12 and reach the peak at nucleus cycle 13, which can reflect the transcription frequency of CG15382. The binding of polymerase decreased in the Zld- mutant.
By searching around the peak of the ChIP result, two Zld binding sites can be found, which are CAGGTAC and CTACCTG. TATA box was found at 30 base pair upstream from the +1 site. The coding region and untranslated region were also determined.
In addition, the binding peak of Zld is not as smooth as other peaks, which might be caused by the convergent of subpeaks.
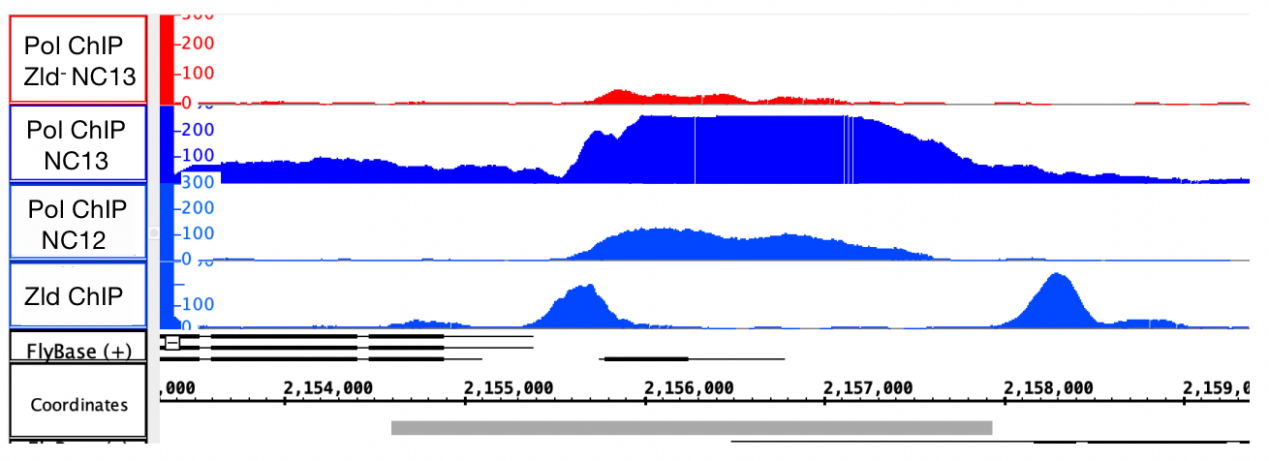
Figure 3. The binding graph of polymerase and Zld protein
The first three rows are the binding graph of polymerase to DNA sequences of individually Zld- mutant at nucleus cycle 13, wild type embryo at nucleus cycle 13 and at nucleus cycle 12. The last row is the binding of Zld protein to wild type embryo DNA. Higher level refers to more combination of protein and DNA sequence.
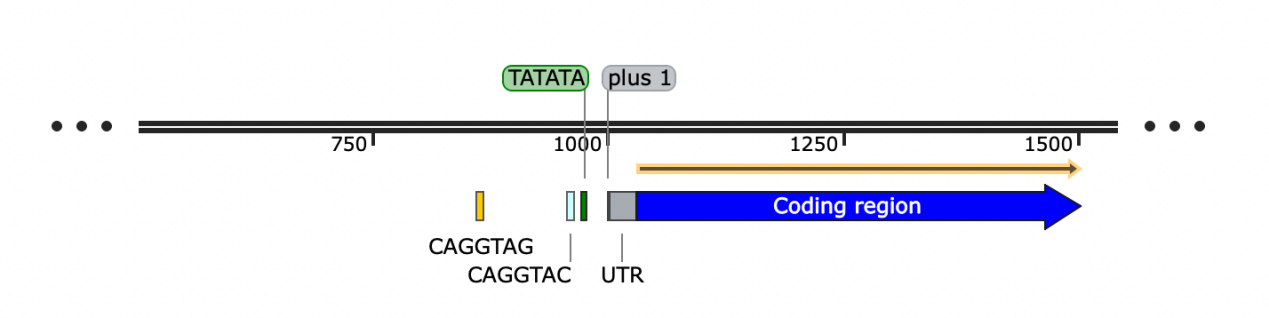
Figure 4. The sketch of CG15382
Plus 1 site is used to locate the sequence. CAGGTAG and CAGGTAC are the two binding site of CG15382. TATATA is the activator sequence of CG15382, which locates at the -30 site. UTR are the abbreviation of untranslated region. The coding region are also marked above.
4.2. CG15382 expression is down-regulated in Zld- mutant
CG15382 was ubiquitously distributed in the embryo. Its expression can be reflected by the fluorescent. By comparing its expression between wild type drosophila and Zld- mutant during different period, it can be found that the expression of CG15382 is down-regulated in Zld- mutant. However, the staining is still evident at the anterior side of the embryo compared to other parts.
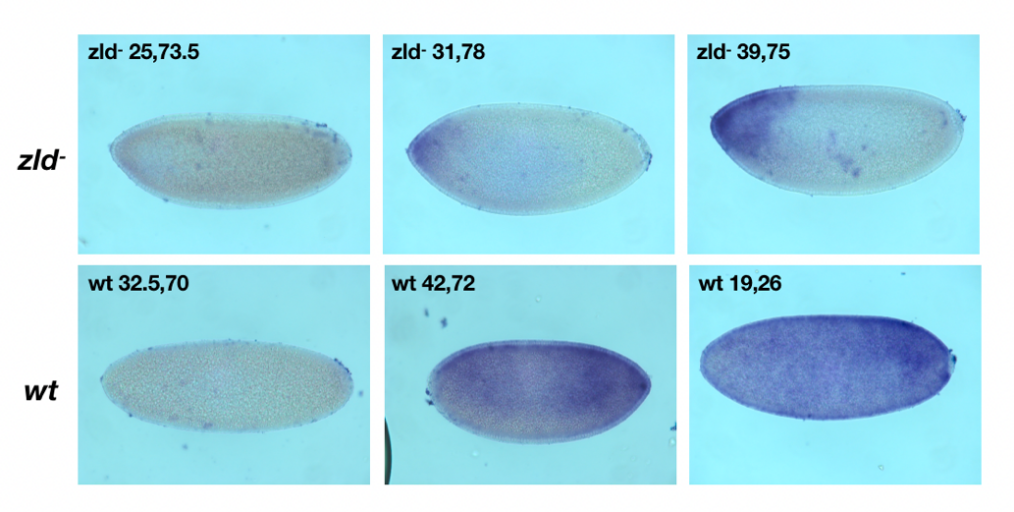
Figure 5. The result of in situ hybridization
The first row is the staining result of CG15382 in zld- mutant at different times. The second row are the result in wild type drosophila at different times.
5. Discussion
According to the result of ChIP, it can be concluded that the binding of polymerase to DNA increases over time and reaches the peak at nucleus cycle 13. However, the binding decreased in the zld- mutant. It might be that lack of zld protein decreases the frequency of DNA replication. In the ChIP of Zld protein, the binding only occurs in a few parts of sequences, and there is a peak around the site of CG15382. However, the peak is not as smooth as others.
By analyzing the sequence in SnapGene software, it was found that there are two binding sequences of Zld protein, which are CAGGTAG and CAGGTAC. That may explain that the peak of Zld is not as smooth: there may be two subpeaks coemerged into one.
To investigate the function of the gene CG15382, gene editing was designed to mutate the binding sequence of its activator Zld. The gene editing was performed by designing a CRISPR-Cas9 system to exchange the binding sequence in CG15382 with CTCATAG. After that, in situ hybridization will be performed (the photos of in situ hybridization was taken from Rushlow unpublished results).
By analyzing the differential expression of Zld between wild type embryos and mutant embryos, it can be concluded that CG15382 is a ubiquitous gene, and its expression level decreases in the case of Zld-. Which shows that the silence of gene CG15382 can be achieved by mutating the binding site of Zld. That may provide a new thought of regulating gene expression.
Besides, the expression of CG15382 in Zld- mutant concentrates around the anterior side of the embryo, which may imply an underlying mechanism of expression regulating. There might be other transcription factors that inhibits the functioning of Zld. Further study can focus on the upstream transcription factor of Zld gene to determine if there are antagonists of Zld protein locates at the anterior part of the early embryo. It is also possible that the CG15382 gene is responsible for regulating the development of the posterior part of Drosophila embryos, and the CG15382 located in the anterior part does not rely on Zld protein activation. Anyway, future experiments can start with the function of CG15382 after comparing and analyzing the differences between the Zld mutation group and the control group.
Overall, the principle of this study is to identify the function of CG15382 by comparing wild type and silenced mutant. The function of genes can be determined by comparing gene silencing with normal groups. It was found that the expression of CG15382 is down regulated in the absence of Zld protein. However, the result of in situ hybridization shows that there are still staining in the anterior part of the Zld- mutant, and the reason remains unknown. As for future study, it can start with the function of gene CG15382 to determine whether it only regulates gene expression at the anterior part. Or it can focus on the upstream gene of Zld to determine if there are Zld protein antagonists present in the anterior part of the embryo.
In addition, the binding affinity can also be investigated. Previous study had identified several binding sites of Zld proteins and determined their respective enrichment indices, however, their binding affinity had not been studied before. The enrichment index of CAGGTAG is the highest among all binding sequences according to previous research, but the peak of CAGGTAC is higher than CAGGTAG in this study [7]. In subsequent studies, the binding affinity of the sequences can be quantified, and whether the difference in binding affinity of different sequences affect the regulation of gene expression can also be investigated.
6. Conclusion
The main finding of this paper is that there are two Zld binding sites in the sequence of CG15382 and its expression is down-regulated in Zld- mutant. It is also suggested that future research can focus on the function of CG15382 and the expression pathway of Zld protein.
References
[1]. Duan J, Rieder L, Colonnetta MM, Huang A, Mckenney M, Watters S, Deshpande G, Jordan W, Fawzi N, Larschan E. CLAMP and Zelda function together to promote Drosophilazygotic genome activation. Elife. 2021 Aug 3;10:e69937. doi: 10.7554/eLife.69937. PMID: 34342574; PMCID: PMC8367384.
[2]. Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008 Nov 20;456(7220):400-3. doi: 10.1038/nature07388. Epub 2008 Oct 19. PMID: 18931655; PMCID: PMC2597674.
[3]. Nien CY, Liang HL, Butcher S, Sun Y, Fu S, Gocha T, Kirov N, Manak JR, Rushlow C. Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet. 2011 Oct;7(10):e1002339. doi: 10.1371/journal.pgen.1002339. Epub 2011 Oct 20. PMID: 22028675; PMCID: PMC3197689.
[4]. Blythe SA, Wieschaus EF. Coordinating Cell Cycle Remodeling with Transcriptional Activation at the Drosophila MBT. Curr Top Dev Biol. 2015;113:113-48. doi: 10.1016/bs.ctdb.2015.06.002. Epub 2015 Jul 29. PMID: 26358872.
[5]. Zhang JP, Li XL, Li GH, Chen W, Arakaki C, Botimer GD, Baylink D, Zhang L, Wen W, Fu YW, Xu J, Chun N, Yuan W, Cheng T, Zhang XB. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 2017 Feb 20;18(1):35. doi: 10.1186/s13059-017-1164-8. PMID: 28219395; PMCID: PMC5319046.
[6]. Ramos JM. Fluorescent In Situ Hybridization (FISH). Methods Mol Biol. 2022;2422:179-189. doi: 10.1007/978-1-0716-1948-3_12. PMID: 34859406.
[7]. ten Bosch JR, Benavides JA, Cline TW. The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development. 2006 May;133(10):1967-77. doi: 10.1242/dev.02373. Epub 2006 Apr 19. PMID: 16624855.
Cite this article
Chang,C. (2024). Research on the function of CG15382 by mutating Zelda binding region. Theoretical and Natural Science,45,47-52.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 2nd International Conference on Modern Medicine and Global Health
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Duan J, Rieder L, Colonnetta MM, Huang A, Mckenney M, Watters S, Deshpande G, Jordan W, Fawzi N, Larschan E. CLAMP and Zelda function together to promote Drosophilazygotic genome activation. Elife. 2021 Aug 3;10:e69937. doi: 10.7554/eLife.69937. PMID: 34342574; PMCID: PMC8367384.
[2]. Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008 Nov 20;456(7220):400-3. doi: 10.1038/nature07388. Epub 2008 Oct 19. PMID: 18931655; PMCID: PMC2597674.
[3]. Nien CY, Liang HL, Butcher S, Sun Y, Fu S, Gocha T, Kirov N, Manak JR, Rushlow C. Temporal coordination of gene networks by Zelda in the early Drosophila embryo. PLoS Genet. 2011 Oct;7(10):e1002339. doi: 10.1371/journal.pgen.1002339. Epub 2011 Oct 20. PMID: 22028675; PMCID: PMC3197689.
[4]. Blythe SA, Wieschaus EF. Coordinating Cell Cycle Remodeling with Transcriptional Activation at the Drosophila MBT. Curr Top Dev Biol. 2015;113:113-48. doi: 10.1016/bs.ctdb.2015.06.002. Epub 2015 Jul 29. PMID: 26358872.
[5]. Zhang JP, Li XL, Li GH, Chen W, Arakaki C, Botimer GD, Baylink D, Zhang L, Wen W, Fu YW, Xu J, Chun N, Yuan W, Cheng T, Zhang XB. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 2017 Feb 20;18(1):35. doi: 10.1186/s13059-017-1164-8. PMID: 28219395; PMCID: PMC5319046.
[6]. Ramos JM. Fluorescent In Situ Hybridization (FISH). Methods Mol Biol. 2022;2422:179-189. doi: 10.1007/978-1-0716-1948-3_12. PMID: 34859406.
[7]. ten Bosch JR, Benavides JA, Cline TW. The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development. 2006 May;133(10):1967-77. doi: 10.1242/dev.02373. Epub 2006 Apr 19. PMID: 16624855.









