1. Introduction
In the exploration of plant growth and development, the role of endogenous hormones is a subject of considerable interest [1]. The experiment is designed to delve into the intricate interactions between plant growth regulators and seed germination, specifically in the model organism Arabidopsis thaliana. Arabidopsis thaliana is widely recognized for its genetic simplicity and rapid lifecycle, making it an exemplary candidate for genetic and physiological studies [2]. It has relatively compact and well-characterized genome which facilitates genetic manipulation and simplifies genomic studies. These features help enabling scientists to identify gene functions and genetic interactions more easily. Additionally, its well-manipulated genes make it an ideal candidate for studying complex traits in genetics. Another significant advantage of Arabidopsis is its short life cycle, which spans only about six weeks from germination to seed maturity. This allows researchers to conduct multiple generations of experiments within a single year, thereby expediting genetic studies and phenotypic analysis. This research leverages the distinct advantages of Arabidopsis to explore the intricate dynamics between plant growth regulators and seed germination, focusing on the effects of two unidentified compounds across three different genotypes of this species. These genotypes presumably include a wild type and two mutants, each with unique responses to hormonal regulation.
Plant hormones, also known as phytohormones. They are crucial in regulating various aspects of plant growth, including the germination process. These organic compounds are produced in extremely low concentrations within plant tissues and orchestrate developmental processes and responses to environmental stimuli. Among the key hormones influencing germination are gibberellins (GAs), abscisic acid (ABA), ethylene, auxins, and cytokinin. Each of them plays a specific and different role in plants development [3]. The interplay among these hormones ensures that the germination occurs at the right time. They also integrate internal developmental cues with external environmental conditions. This hormonal balance not only triggers the start of germination but also coordinates subsequent growth stages. Thereby optimizing the plant’s chances for survival and successful growth. The rationale behind this experiment is to better understand how different plant growth regulators can influence germination. Previous studies have demonstrated that GA and ABA antagonistically regulate seed maturation, hypocotyl, and stem elongation [4]. Gibberellins are vital for initiating germination [5]. They promote the breakdown of seed dormancy and facilitate the emergence of the radicle by stimulating the production of enzymes that degrade the starch stored in the seed endosperm into glucose. This process provides the necessary energy for the developing embryo. Grasping the mechanisms of GA transport is vital for the survival of plant species and the success of crop production [6]. Abscisic Acid maintains seed dormancy and acts as a growth inhibitor by suppressing the action of gibberellins. High levels of ABA prevent germination, making its downregulation for the seed to sprout. The dynamic balance between ABA and gibberellins is therefore critical for ensuring that seeds germinate only under favourable conditions. Yet, mutations resulting in inadequate ABA levels cause significant stunting in plants [7]. Studying seed germination under the influence of these regulatory substances can provide valuable understanding of the genetic mechanisms that control plant growth reactions.
This study aims to dissect the germination rates across the selected Arabidopsis genotypes under the influence of these regulatory substances to elucidate potential genetic mechanisms underlying plant growth responses. This study is predicated on three null hypotheses. The first null hypothesis states that the genotype of Arabidopsis, whether G1, G2, or G3, does not significantly influence the rate of seed germination, indicate that any observed differences in germination rates are due to random chance rather than genotype. The second null hypothesis suggests that the treatment applied to the seeds (exposure to ethanol, Compound A, or Compound B) has no significant impact on the germination rates, thereby implying that all treatments are equally effective in influencing seed germination. The third null hypothesis assumes that there is no significant interaction between the genotype of the Arabidopsis seeds and the treatments applied; in other words, the effect of the treatment on germination rates is uniform across all genotypes, without any genotype responding differently to the treatments.
The experiment investigates the varying germination rates across genotypes and seeks to associate these rates with possible genetic variations that affect their reactions to the compounds. It also validates the comprehension of hormone-regulated germination, and potentially uncover information about the genotypes’ identities through their phenotypic reactions.
2. Materials and method
2.1. Reagents
Murashige and Skoog Basal Salt Mixture (MS), agar, and milliQ water. Prepared 2x agar and 2x MS: one with 1.6g of agar and another with 0.44g of MS salts, each diluted in 100 mL of water to achieve a 2x concentration. These solutions were autoclaved to ensure sterility.
2.2. Planting medium preparation
10 µM of Compound A and 3 µM of Compound B were added into separate aliquots of the 2x MS solution, along with a mock treatment using ethanol as a control. Each treatment was then mixed with an equal volume of 2x agar to form a 1x planting medium.
2.3. Seeds incubation
The media was dispensed into labelled Petri dishes, dividing each into three sectors to allocate space for each of the three Arabidopsis genotypes: G1, G2, and G3. Approximately 30 seeds from each genotype were then evenly distributed into their respective sectors using a sterilized aluminium sheet to facilitate precise placement and avoid cross-contamination. After a stratification period of four days at 4°C and subsequent incubation under light for seven days to simulate natural germination conditions, then assessed germination by counting the emerged seedlings.
2.4. Germination rate calculation
Germination rate calculation was performed by using dissecting microscopes to ensure accuracy, count total number of seeds in each sector. This number represented the seeds that germinated. Seedlings have come out of the seed coat and have green cotyledons. Calculated germination percentage for each genotype under different conditions by using this formula: Number of seeds that germinated) / (Total number of seeds) *100.
2.5. Statistical data analysis
The raw data were collected the germination percentage for each genotype under each treatment condition were calculated. These data were plotted in individual graphs to visually represent the germination responses. Statistical analyses, including two-way ANOVA, were conducted to determine if the differences observed across genotypes and treatments were significant, thereby testing our initial hypotheses regarding the effect of the genotypes and the impact of the compounds on germination rates.
3. Results
3.1. Genotype-specific responses to the various media
Visual analysis of germination patterns presented in Figure 1 revealed genotype-specific responses to the various media. G1 exhibited consistent germination across ethanol and Compound A treatments but showed inhibited germination with Compound B. G2 germination was unaffected by Compound B, while G3 showed poor germination unless exposed to Compound A.
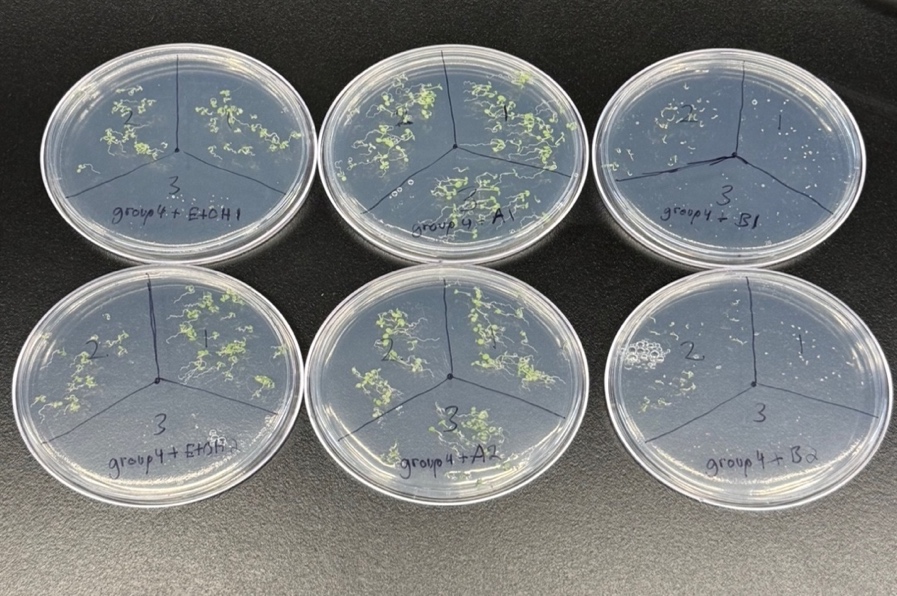
Figure 1. Germination of Different Arabidopsis Genotypes on Different Media. This figure displays Petri dishes containing Arabidopsis seeds from three unknown genotypes (labeled as 1, 2, and 3) after a germination period. Each plate is divided into three sections, each containing one genotype. The media in the plates include a control (ethanol), compound A, compound B, labeled as EtOH, A, and B. All the media have replicate A plate and replicate B plate.
3.2. Responses of different genotype with treatments
Results showed that genotype G1 exhibited optimal germination in the ethanol and Compound A media, achieving a 100% success rate, yet it failed to germinate in the Compound B medium. Genotype G2 also reached a 100% germination rate in both the ethanol and Compound A media, with a slightly reduced rate of approximately 87.5%, 56.67%, 79.3%, and 100% in the Compound B medium. Genotype G3’s response was disparate, with complete germination on the Compound A medium, whereas its success rates on ethanol and Compound B media were markedly lower, at 9.38%, 5.88%, 0%, and 2.7% on ethanol; 0% on Compound B media (Figure 2).
Figure 2. Germination Response of Different Arabidopsis Genotypes to Ethanol, Compound A, Compound B. This bar chart illustrates the germination percentages of three Arabidopsis genotypes (G1, G2, and G3) when treated with ethanol (EtOH), Compound A, and Compound B. Each genotype’s response is represented by three bars corresponding to the different treatments.
3.3. Germination rates of different genotype with treatments
As Figure 3 showed that genotype G1 and G2 had complete germination in the ethanol and Compound A media. However, G1 had no germination in the Compound B medium. Genotype G2 had 100% germination rates on both ethanol media and Compound A media, while its mean germination rate decreased to 80.87% on Compound B media. Genotype G3 had a low germination rate 4.49% in ethanol, but it achieved full germination in Compound A. Yet, it showed no germination in the Compound B medium, much like G1. These variations in germination rates across the different treatments revealed the unique interactions between each genotype and the growth media.
Figure 3. Mean Germination Rates of Arabidopsis Genotypes Across Different Media. This bar graph displays the mean germination rates of three Arabidopsis thaliana genotypes (G1, G2, and G3) exposed to different treatments: ethanol (EtOH), Compound A, and Compound B.
The statistical analysis indicated significant differences in germination rates as shown in Table 1 by conducting a Two-way ANOVA test. Genotype had a significant effect on the dependent variable, F(2, 27) = 267.96902, p = 1.55666 x 10^-18. The media type also significantly influenced the germination outcome, F(2, 27) = 414.94574, p = 5.35688 x 10^-21. Additionally, there was a significant interaction effect between genotype and media, F(4, 27) = 135.21291, p = 1.91842 x 10^-17.
Table 1. Two – way ANOVA of Arabidopsis Genotypes Grown on Different Media
Source of Variance | ss | df | MS | F | P-value | F crit |
Genotype | 20786.9934 | 2 | 10393.497 | 267.96902 | 1.5567E-18 | 3.3541308 |
Media | 32188.3267 | 2 | 16094.163 | 414.94574 | 5.3569E-21 | 3.3541308 |
Interaction Genotype * Media | 20977.5737 | 4 | 5244.3934 | 135.21291 | 1.9184E-17 | 2.7277653 |
Within | 1047.22708 | 27 | 38.786188 | |||
Total | 75000.121 | 35 |
As the Tukey test resulted in Table 2, significant differences in germination rates were observed between all genotypes, with G1 versus G2, G1 versus G3, and G2 versus G3 all yielding ‘yes’ for significance (abs dif of means > 5.447). Comparisons between treatments also showed significant differences (abs dif of means >5.447), particularly between ethanol versus Compound A, Compound A versus Compound B, and ethanol versus Compound B. Further, within-genotype comparisons confirmed that G1’s germination was unaffected by ethanol and Compound A (abs dif of means <17.108) but was significantly inhibited by Compound B (abs dif of means >17.108). G2 germination rates did not vary between ethanol and Compound A (abs dif of means <17.108) but significantly showed variance with Compound B (abs dif of means >17.108). While G3 had significant different germination rates compared the Compound A media with ethanol or Compound B respectively (abs dif of means >17.108), no significant differences with the comparison between ethanol and Compound B media (abs dif of means <17.108).
Table 2. Tukey Post-Hoc Comparisons of Arabidopsis Genotypes and Treatment Effects on Germination
Comparison | Abs Dif of Means | Crit Value | Significant | df | k | n | MSwithin | q value |
EtOH VS Compound A | 31.8366667 | 5.447416454 | Yes | 27 | 3 | 12 | 38.786188 | 3.03 |
Compound A VS Compound B | 73.0433333 | 5.447416454 | Yes | |||||
G1& Compound A VS G1& Compound B | 41.2066666 | 5.447416454 | Yes | |||||
Comparison | Abs Dif of Means | Crit Value | Significant | df | k | n | MSwithin | q value |
G1& EtOH VS G1& Compound A | 0 | 17.1081 | no | 27 | 9 | 3 | 38.786188 | 4.758 |
G1& EtOH VS G1 & Compound B | 100 | 17.1081 | yes | |||||
G1& Compound A VS G1& Compound B | 100 | 17.1081 | yes | |||||
G2& EtOH VS G2& Compound A | 0 | 17.1081 | no | |||||
G2& EtOH VS G2 & Compound B | 19.13 | 17.1081 | yes | |||||
G2& Compound A VS G2& Compound B | 19.13 | 17.1081 | yes | |||||
G3 & EtOH VS G3 & Compound A | 95.51 | 17.1081 | yes | |||||
G3& EtOH VS G3 & Compound B | 4.49 | 17.1081 | no | |||||
G3& Compound A VS G3 & Compound B | 100 | 17.1081 | yes |
4. Discussion
Upon thorough analysis of the experimental data, we have found compelling evidence to reject all three of the null hypotheses presented in our study. The first null hypothesis was rejected by statistically significant differences in germination observed among the genotypes (p < 0.05). This indicates that genetic variation among these genotypes has a determinable effect on their ability to germinate under controlled conditions [8]. However, the data presented clear disparities in germination outcomes dependent on the compounds applied (p < 0.05), leading us to reject the second hypothesis as well. The distinct germination patterns indicate that these compounds have specific roles and efficacies in influencing seed germination [9]. Lastly, contrary to the third null hypothesis, our results demonstrated a significant interaction (p < 0.05), revealing that genotypes responded differently to each treatment. This suggests that Arabidopsis seeds’ genotypic makeup influences how they react to the applied growth regulators, demonstrating that the genotype-treatment interaction is a key factor in the germination process [10].
Based on the germination rates observed, Compound A could be hypothesized to be a growth regulator that promotes germination, such as gibberellic acid, because it enhances germination in G3, which otherwise shows poor germination. Compound B may be an inhibitor of germination, such as abscisic acid, since both G1 and G3 exhibit reduced or no germination with it. The complete germination inhibition confirms that Compound B may activate a germination-inhibitory pathway or possess toxicity to which the seeds cannot respond [11].
If one of the unknown compounds is a bioactive gibberellin and the other is an ABA inhibitor, genotype G1 is likely the wild type because it shows a high and consistent germination rate across Ethanol and Compound A media, and its germination rate is expected to decrease with ABA treatment due to the compound’s known role as a growth inhibitor. The G2 genotype hypothesized to a mutant that is insensitive to ABA, because it only exhibited a decrease in germination in the presence of ABA compare with the wild-type response [12]. G2 genotype’s reaction to ethanol should mirror that of G1, and one would expect a normal increase in germination rates in response to GA. Lastly, the G3 genotype which supposedly lacks the ability to produce its own GA, presented with a low germination rate in ethanol, as no external GA was provided to facilitate the process [13]. When exposed to GA, it is predicted that G3 would exhibit a germination response approaching that of the wild type, as the externally supplied GA should compensate for its inherent deficiency. The addition of ABA would likely exacerbate this effect, further inhibiting germination in the third plate. Future experiments could assess whether the sensitivity or insensitivity to gibberellic acid and abscisic acid in plants correlates with tolerance to other stress factors such as drought or salinity, contributing to broader agronomic implications.
5. Conclusion
In conclusion, the results are consistent with established scientific knowledge that gibberellic acid generally promotes germination, whereas abscisic acid inhibits germination. The collective evidence from experiment leads to a confident identification of Compound A as gibberellic acid and Compound B as abscisic acid. Genotypic responses to these compounds are as expected: G1 as wild-type, G2 as ABA-insensitive, and G3 as GA-deficient, thus confirming the alternative hypothesis with rationale supported by logic and data. These findings enhance the understanding of genotype-specific responses to plant growth regulators and offer valuable insights into the genetic control of seed germination.
References
[1]. Mukherjee, A.; Gauray, A.K.; Singh, S.;Yadv, S.; Bhowmick, S.; Abeysinghe, S.; Verme, J.P. (2022). The bioactive potential of phytohormones: A review. Biotechnol Rep (Amst) 35, e00748.
[2]. Koornneef, M., & Meinke, D. (2010). The development of Arabidopsis as a model plant. The Plant journal : for cell and molecular biology, 61(6), 909–921. https://doi.org/10.1111/j.1365-313X.2009.04086.x.
[3]. Liu,X.; Hou,XL. (2018). Antagonistic Regulation of ABA and GA in Metabolism and Signaling Pathways. Front Plant Sci 9,251.
[4]. Shu, K.; Zhou, W.; Chen, F.; Luo, X.; Yang, W. (2018). Abscisic Acid and Gibberellins Antagonistically Mediate Plant Development and Abiotic Stress Responses. Front Plant Sci 9, 416.
[5]. Weiss, D.; Ori, N.(2007). Mechanisms of Cross Talk between Gibberellin and Other Hormones. Plant Physiol, 144(3):1240-1246.
[6]. Gupta,R.; Chakrabarty, S.K.;(2013). Gibberellic acid in plant. Plant Signal Behav 8(9):e25504.
[7]. Finkelstein, R. Abscisic Acid Synthesis and Response. Arabidopsis Book 11:e0166.
[8]. Geshnizjani, N.; Snoek,B.; Willems, L.; Bienstra, J.; Nijveen, H.; Hilhorst, H.; Ligterink, W. (2022). Detection of QTLs for genotye * environment interactions in tomato seeds and seedlings. Plant Cell Environ, 43(8):1973-1988.
[9]. Zhao, H.; Zhang, Y.; Zheng, Y.; Integration of ABA, GA, and light signaling in seed germination through the regulation of ABI5. (2022). Front Plant Sci. 13:1000803.
[10]. Castaño G.; Cabrera.C.; Pernas, M.; Gómez, L.; Sánchez,L.; An updated overview on the Regulation of Seed Germination. (2020). Plants. 9(6):703.
[11]. Finkelstein, R.R.; Gampala, S.S.L.; Rock, C.D. (2002). Abscisic Acid Signaling in Seeds and Seedlings. Plant Cell 14, s15-s45).
[12]. Finkelstein, R.R.; Somerville, C.R.(1990). Three classes of Abscisic Acid – Insensitive Mutations of Arabidopsis Define Genes that Control Overlapping Subsets of ABA Response. Plant Physiol, 94(3):1172-1179.
[13]. Peng,J.; Harberd, N.P. (1997). Gibberellin deficiency and response mutations suppress the stem elongation phenotype of phytochrome-deficient mutants of Arabidopsis. Plant Physiol 113(4);1151-1058.
Cite this article
Xu,X. (2024). Gibberellic Acid and Abscisic Acid Effects on Germination Across Arabidopsis Genotypes: Confirmation of Compound Identity and Genotypic Responses. Theoretical and Natural Science,54,55-62.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of ICBioMed 2024 Workshop: Workshop on Intelligent Medical Data Analysis for Precision Medicine
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Mukherjee, A.; Gauray, A.K.; Singh, S.;Yadv, S.; Bhowmick, S.; Abeysinghe, S.; Verme, J.P. (2022). The bioactive potential of phytohormones: A review. Biotechnol Rep (Amst) 35, e00748.
[2]. Koornneef, M., & Meinke, D. (2010). The development of Arabidopsis as a model plant. The Plant journal : for cell and molecular biology, 61(6), 909–921. https://doi.org/10.1111/j.1365-313X.2009.04086.x.
[3]. Liu,X.; Hou,XL. (2018). Antagonistic Regulation of ABA and GA in Metabolism and Signaling Pathways. Front Plant Sci 9,251.
[4]. Shu, K.; Zhou, W.; Chen, F.; Luo, X.; Yang, W. (2018). Abscisic Acid and Gibberellins Antagonistically Mediate Plant Development and Abiotic Stress Responses. Front Plant Sci 9, 416.
[5]. Weiss, D.; Ori, N.(2007). Mechanisms of Cross Talk between Gibberellin and Other Hormones. Plant Physiol, 144(3):1240-1246.
[6]. Gupta,R.; Chakrabarty, S.K.;(2013). Gibberellic acid in plant. Plant Signal Behav 8(9):e25504.
[7]. Finkelstein, R. Abscisic Acid Synthesis and Response. Arabidopsis Book 11:e0166.
[8]. Geshnizjani, N.; Snoek,B.; Willems, L.; Bienstra, J.; Nijveen, H.; Hilhorst, H.; Ligterink, W. (2022). Detection of QTLs for genotye * environment interactions in tomato seeds and seedlings. Plant Cell Environ, 43(8):1973-1988.
[9]. Zhao, H.; Zhang, Y.; Zheng, Y.; Integration of ABA, GA, and light signaling in seed germination through the regulation of ABI5. (2022). Front Plant Sci. 13:1000803.
[10]. Castaño G.; Cabrera.C.; Pernas, M.; Gómez, L.; Sánchez,L.; An updated overview on the Regulation of Seed Germination. (2020). Plants. 9(6):703.
[11]. Finkelstein, R.R.; Gampala, S.S.L.; Rock, C.D. (2002). Abscisic Acid Signaling in Seeds and Seedlings. Plant Cell 14, s15-s45).
[12]. Finkelstein, R.R.; Somerville, C.R.(1990). Three classes of Abscisic Acid – Insensitive Mutations of Arabidopsis Define Genes that Control Overlapping Subsets of ABA Response. Plant Physiol, 94(3):1172-1179.
[13]. Peng,J.; Harberd, N.P. (1997). Gibberellin deficiency and response mutations suppress the stem elongation phenotype of phytochrome-deficient mutants of Arabidopsis. Plant Physiol 113(4);1151-1058.









