1. Introduction
Genome editing is an advanced technique in genetic manipulation, involving the deliberate alteration of DNA within an organism's genome. This process typically involves the precise introduction of a double strand break (DSB) at a targeted location within the DNA sequence. To facilitate this targeted DNA modification, scientists have utilized engineered nucleases, often referred to as "molecular scissors," which are designed to cut DNA at specific sites. These tools enable researchers to insert, remove, or replace DNA segments with high accuracy, thereby allowing for the manipulation of genetic material in a controlled manner. Artificial endonuclease-mediated genome editing technologies include four main types: meganuclease, ZFN, TALEN, and CRISPR/Cas system [1,2]. ZFNs were pioneering tools for DNA cleavage, allowing for targeted genome editing. However, they had a notable drawback: a tendency to cause off-target mutations, which could lead to unintended genetic changes [3]. TALENs are large, complex genome-editing proteins that can be difficult to deliver efficiently using viral vectors due to their size and repetitive regions [4]. Compared with ZFN and TALENs gene editing technology, The CRISPR/Cas system is valued for its affordability, effectiveness, and wide application, thus being used in various scientific and medical fields. This system's accessibility and reliability have broadened its adoption in genetic research and therapeutic applications.
Originally CRISPR/Cas system is found in bacteria, serves as an adaptive immune system that enables them to efficiently recognize and cleave invading exogenous nucleic acids. Back to the 1980s, derived from studies of bacteria and archaea, scientists discovered a special type of repeating sequence, known as the CRISPR sequence. In 2007, scientists first experimentally showcased that the CRISPR system can act as an acquired immune mechanism for bacteria to defend against the invasion of foreign genetic material. CRISPR technology's swift advancement is transforming scientific research, enabling genetic manipulation in model organisms and altering the traits of key agricultural species. Additionally, it holds promise for medical applications, particularly in addressing genetic disorders. CRISPR genome editing has reached a milestone with the initial regulatory approval of therapies based on this technology for treating human diseases in late 2023 [5]. This marks the beginning of a transformative period in medical treatment.
2. Classification of CRISPR/Cas system
According to the Cas protein of which it is composed, CRISPR/Cas systems are divided into two primary classes, each with a distinct set of characteristics and mechanisms [6].
2.1. Class I CRISPR/Cas systems
These CRISPR/Cas systems encompass intricate effector complexes composed of multiple Cas proteins. The first class of CRISPR/Cas systems is further segmented into three categories:
First is Type I, at the heart of which is the Cas3 protein, a member of the helicase family that has the ability to unravel DNA-DNA and RNA-DNA double strands. When cutting DNA, the Cas3 complex of a Class I CRISPR system typically moves along the DNA strand, causing cuts at multiple sites. Type III systems are similar to Type I systems, but they use fewer Cas proteins to perform genome editing tasks. Type III systems also require crRNA to direct the Cas protein complex to the target DNA, but they do not require tracrRNA. Cas10 is a key protein in the Type III system, responsible for recognizing PAM (the site adjacent to the original interspaced short palindromic repeat) and cutting DNA. Type IV, a rarer type of CRISPR/Cas system that is less studied, has been known that they also involve multiprotein complexes to perform gene editing functions. The complexity of Class I systems provides them with more diverse editing capabilities and application potential, although it also means that they may face more technical challenges in practical applications. Scientists are exploring the potential of utilizing the Class I CRISPR/Cas system as a means to activate transcription in human cells [7].
2.2. Class II CRISPR/Cas systems
These systems consist of effector complexes of single Cas proteins, which also include three types. As the most widely researched and used system, Type II system (Figure 1). The Cas9 protein s integral to the procedure, using the guide RNA to recognize specific PAM sequences and cut double-stranded DNA in the immediate vicinity of the PAM. Cas proteins in Type V, such as Cas12a (previously known as Cpf1) [8], are used for RNA targeting and DNA editing. Cas12a does not require tracrRNA, but instead uses crRNA to guide its targeting. The cutting activity of Cas12a can be used for multiple edits, and they have different requirements for PAM sequences than Cas9. The last Type VI utilizes Cas13 proteins, which are primarily targeted at RNA editing. Cas13 proteins can use a specific crRNA to identify and cleave target RNA molecules, providing new tools for RNA-directed gene silencing and research. Owing to its user-friendly nature and remarkable editing proficiency, and relatively fewer off-target cuts, Type II systems, especially CRISPR/Cas9 system, have become the tool of choice for many gene-editing experiments.
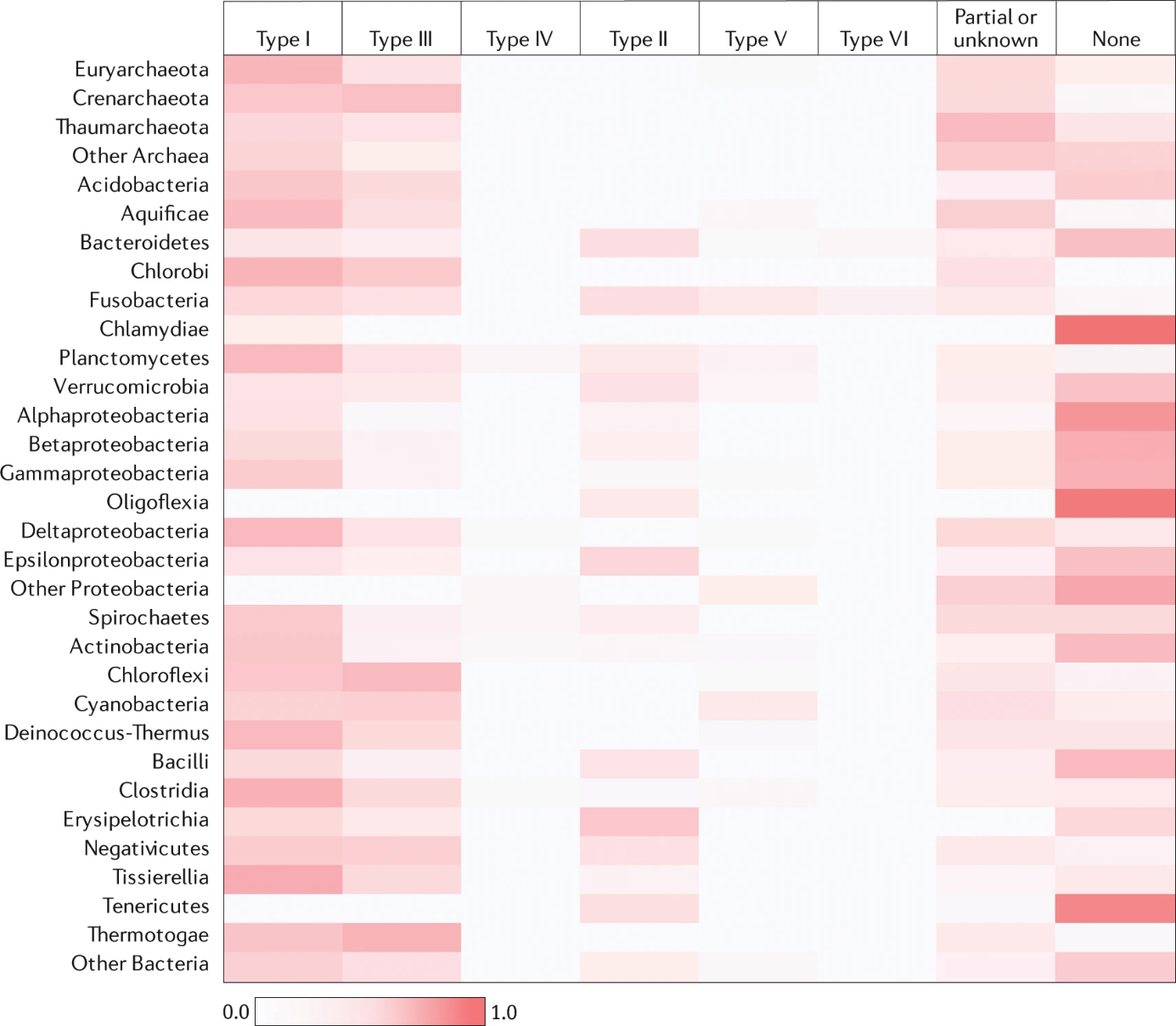
Figure 1. Distribution of six types of CRISPR-Cas system in the archaeal and bacterial phyla [8].
3. Molecular mechanisms of the CRISPR/Cas system
The CRISPR/Cas9 system operates through a sequence of distinct stages: adaptation, expression and interference.
During the adaptation stage, bacteria recognize and cut invading exogenous DNA (called protospacer) via Cas proteins, and then integrate these DNA fragments into CRISPR sequences as new spacer sequences. Subsequent to the expression phase, the CRISPR sequences undergo transcription to form the precursor RNA (pre-crRNA), and then the pre-crRNA is refined into mature crRNA (CRISPR RNA) by the action of RNase III and other nucleases. These mature crRNA molecules contain sequences complementary to previously captured viral DNA sequences. In the final phase of interference, the mature crRNA binds to the Cas9 protein to form an RNA-protein complex that searches the bacterial cell for foreign DNA sequences complementary to the crRNA and directs the Cas9 protein to make cuts that can damage the DNA of the invading virus or plasmid. The cuts usually occur near a specific Protospacer Adjacent Motif (PAM) sequence, which is located upstream of the potential cleavage site, guides the Cas protein to correctly localize to the target site and cleave. Each variant of Cas proteins is capable of identifying and responding to distinct PAM sequences. For example, the most widely used SpCas9, originating from Streptococcus pyogenes, is known for its specificity to the PAM sequence 5'-NGG-3', where N stands for any nucleotide and G is an essential nucleotide.
The initial iteration of CRISPR technology predominantly relies on intrinsic repair mechanisms or precise DNA DSBs. Developed to the second generation CRISPR technology, it can mediate genome editing without relying on DNA double-strand break formation and HDR, the most representative of which are base editor (BE) and prime editor (PE) [9]. Base Editors mainly include Cytosine Base Editors (CBEs) and Adenine Base Editors (ABEs), both of which are designed to convert a base at a target site to another base by a specific deaminase enzyme, thus enabling base substitutions. Prime Editor is even more advanced, allowing virtually any of the 12 required individual base substitutions, minor insertions or omissions to be fitted to a specific location in the genome. The latest developments show that PM359, the first therapeutic candidate of the Prime Editor technology, has received clearance from the US Food and Drug Administration for addressing Chronic Granulomatous Disease (CGD) in a Phase 1/2 clinical experiment, and the publication of data from the preliminary clinical trial is expected in 2025, which facilitates an evaluation of the viability of employing this technology.
4. Applications of CRIPSR/Cas system in genetic diseases
4.1. Hemoglobinopathies
Both of Beta-thalassemia and sickle cell disease are caused by mutations in the hemoglobin gene, resulting in abnormal hemoglobin functioning. In sickle cell patients, there is a single small mutation in the gene that codes for hemoglobin. Valine replaces glutamate in the sixth amino acid position. In erythrocytes, when HBS loses the oxygen it carries, it polymerizes with itself, forming chain-like structures that deform the cell [10,11].
Casgevy can initiate another type of hemoglobin encoded in our genes -- fetal hemoglobin (HBF) -- and when HBF is mixed with HBS, it prevents the mutant protein from polymerizing itself, then HBS fails to form the structure that deforms red blood cells.
Casgevy leverages CRISPR/Cas9 to enhance the production of fetal hemoglobin in stem cells. This fetal hemoglobin, present in limited quantities in adults but abundant in fetuses, can substitute for the deficient adult hemoglobin. When these engineered stem cells are administered to patients, they can elevate red blood cell counts in those with beta-thalassemia and mitigate the painful episodes experienced by sickle cell disease patients [12]. Before the FDA, a Casgevy approved clinical trial in the U.K. showed that 28 of the 29 patients who participated in the trial were free of severe pain within 12 months of treatment in the trial for SCD. Specifically, in certain adults, the mutation of the BCL11A gene has been linked to the production of fetal hemoglobin. Casgevy replicates this effect by utilizing gene-editing techniques to precisely target and inactivate the BCL11A gene within hematopoietic stem cells. This intervention aims to stimulate the release of fetal hemoglobin, offering a novel therapeutic avenue for various blood disorders.
4.2. Ocular disease
The CRISPR system demonstrates significant promise in addressing Leber congenital amaurosis (LCA). LCA is an inherited disorder affecting the retina, marked by the gradual deterioration of photoreceptor cells and the retinal pigment epithelium (RPE) [13], usually resulting from mutations in genes such as CEP290, leading to vision loss and retinal degeneration. The CRISPR/Cas9 system has been employed to construct a cellular model that simulates LCA10, and by using specific sgRNAs and a combination of Cas9, the IVS26 mutation in the CEP290 gene has been corrected, effectively restoring the mutation in the LCA cellular model. In a clinical trial, a total of 12 adult and two child participate in a gene editing therapy for one eye from 2020 to 2023.
Glaucoma is an ocular disease defined by increased intraocular pressure with damage to the optic nerve that can lead to irreversible vision loss. Recent studies have found that targeting the knockout of the carbonic anhydrase 2 (Car2) gene, which can sustain intraocular pressure by increasing aqueous humor production, through CRISPR-Cas9 technology could achieve a sustained reduction in intraocular pressure for more than two months after a single dose in a mouse model [14]. In comparison with the carbonic anhydrase inhibitors (CAIs) that are typically utilized in the management of glaucoma, the targeted ablation of the Car2 gene has exhibited enhanced therapeutic benefits, particularly in its ability to significantly lower intraocular pressure and impede the advancement of the disease.
Inherited retinal diseases, often abbreviated as IRDs, represent a diverse array of conditions that lead to vision loss due to the genetic factors involved. These disorders are typified by the gradual deterioration of the photoreceptor cells and the retinal pigment epithelium, which are critical components of the retina responsible for maintaining visual function. The most common type is Retinitis pigmentosa (RP), with no efficiency therapy currently. One study employed an adenine base editor to repair curative mutations in an RP mouse model by making precise changes to specific base pairs within the DNA sequence without cutting the DNA and delivered using a dual AAV vector, thereby reducing the risk of creating unintended mutations [15]. Through a single subretinal injection, adenine base editors delivered by double AAV precisely corrected pathogenic single nucleotide mutations located in retinal nerve cells with an effect of up to 49%, and the subsequent behavioral results also showed that the mice had improved visual acuity.
4.3. Muscular genetic disease
Duchenne muscular dystrophy (DMD) is an inherited disease stemming from alterations in the DMD gene that results in muscle cells failing to properly express the dystrophin protein (dystrophin), which causes muscle degeneration. In a particular research project, investigators harnessed the potential of a fibrin-based scaffold to cultivate muscle stem cells (MuSCs) derived from unprocessed muscle tissues of mdx mice, a naturally occurring mutation with a C-to-T transition mutation in the 23rd exon that results in premature codon termination [16]. which serve as a representative model for DMD. By employing the precision of CRISPR/Cas9 genome-editing technology, they were able to rectify a mutation associated with myotonic dystrophy proteins within these proliferated MuSCs. Subsequently, the reintroduction of these corrected cells into the mdx mice's skeletal muscle tissue resulted in the reinstatement of the myotonic dystrophy protein expression [17,18]. CRISPR-mediated exon excision can act as a tool for correcting exon deletion mutations in the antiatrophic protein gene, and deletion of the exon 45-55 hotspot segment treats more than 60% of DMD patients [19]. However, in October 2022, the world's only volunteer with DMD who participated in CRISPR gene editing therapy died during the treatment, which suggested the treatment needed further research.
Facioscapulohumeral Muscular Dystrophy (FSHD) , a genetic disorder affecting muscles, is noted for its gradual deterioration of muscle strength and the subsequent wasting away of muscle tissue. Some of the types may also involve the heart, skeletal system. FSHD ranks among the frequently occurring progressive muscle diseases, influencing at least 500,000 people worldwide. Approximately two-thirds of cases are inherited from parents. One-third of the disease appears to be the result of a spontaneous genetic mutation. Currently, FSHD is thought to be caused by a gene called DUX4. The most direct way to treat FSHD is to inhibit the DUX4 gene or modify it to eliminate its activity. Numerous copies, either identical or very similar of the DUX4 are interspersed in repetitive DNA sequences throughout the human genome (called D4Z4). Therefore, rather than using the more traditional CRISPR/Cas9 system to silence the DUX4 gene, the researchers are developing another system based on the CRISPR Cas13 protease, which targets the dux4 mRNA instead of the DNA. to achieve DUX4 gene silencing, but without disrupting the genome in potentially hundreds of places [20].
4.4. Genetic liver disease
Applications in the treatment of transthyretin amyloidosis (ATTR) have been primarily seen in the development of the CRISPR/Cas9-based gene editing therapy NTLA-2001.This therapy delivers mRNA sequences carrying sgRNAs targeting disease-causing genes and optimized SpCas9 proteins to the liver utilizing a lipid nanoparticle delivery vector designed to alleviate ATTR amyloidosis by lowering the serum concentration of transthyretin (TTR) protein to treat ATTR amyloidosis. This toxic TTR protein builds up on nerves and the heart, triggering pain, numbness and heart disease. Positive results were achieved in Phase I clinical trials: a single dose of NTLA-2001 resulted in an average 87% reduction in serum transthyretin protein (TTR) levels, with a maximum of 96%.
The condition known as Alpha-1 Antitrypsin Deficiency (AATD) is a result of genetic variations in the SERPINA1 gene, which induces the alpha-1 antitrypsin protein to fold abnormally and accumulate in the liver, causing liver damage and lung dysfunction. CRISPR/Cas9 gene editing technology served to correct mutations in genes that cause AAT deficiency, and targeted gene correction was successfully performed in the livers of AATD model mice to restore low levels of normal AAT to normal levels.
4.5. Potential challenges of CRISPR technology in disease treatment
The CRISPR system may produce cuts in non-targeted regions during gene editing, leading to unexpected mutations, which is known as off-target effects. Therefore, scientists use guide RNA (sgRNA), which targets Cas nuclease to a precise site in the mutated gene. However, this leads to inefficiencies in gene editing and increases the cost of its use.
Delivery methods are also a major challenge in the development of CRISPR technology, particularly for organs apart from the hematological and hepatic systems, and for certain in vitro cell types. Currently AAV stands out as the most prevalent delivery method for the application of CRISPR/Cas technology in vivo. Viral vectors can efficiently deliver Cas9 genes or Cas9 mRNA into cells, but may be accompanied by risks of immunogenicity and genomic integration.
5. Conclusion
As CRISPR technology continues to advance, its capability in the treatment of human diseases is becoming increasingly apparent. This article reviews the fundamentals of the CRISPR/Cas system, its applications in genetic diseases, and its progress in clinical trials. CRISPR technology, distinguished by its accurate gene editing abilities, offers new hope for the treatment of diseases that are difficult to address by traditional methods. With its precise gene editing capabilities, the advanced technique provides promising prospects for the treatment of diseases that are difficult to address by traditional methods. Despite the tremendous therapeutic potential that CRISPR technology has demonstrated, it must be recognized the challenges that it poses in terms of safety, ethics, and accessibility. Off-target effects of gene editing, long-term impacts, and potential effects on the gene pool all require further research and assessment. With the maturation of the technology, interdisciplinary cooperation, and the improvement of management systems, CRISPR technology will play an even greater role in the medical field, bringing about significant changes.
References
[1]. Townsend JA, Wright DA, Winfrey RJ,et al. High-frequency modification of plant genes sing engineered zinc-finger nucleases[J]. Nature, 2009, 21;459(7245):442-5.
[2]. Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing[J]. at Rev Mol Cell Biol, 2013,14(1):49-55.
[3]. Wang Linlin, Li Hongling. Application of CRISPR/Cas9 gene editing technology in precision ncology research[J].Chinese Journal of Cancer Biotherapy, 2024,31(05):519-527.
[4]. Xue Shan, Wang Shuya, Liu Li, et al. Research progress on precision gene editing technology ased on CRISPR/Cas9[J].Chinese Journal of Biotechnology, 2023,39(07):2566-2578.
[5]. Wong, C. UK first to approve CRISPR treatment for diseases: what you need to know[J]. ature, 2023, 623, 676–677.
[6]. Xin Yuan, Tian Kairen, Qiao Jianjun, et al. Gene editing tools based on CRISPR/Cas system nd their improvement strategies[J].Chinese Journal of Biotechnology, 2023,43(08):72-85.
[7]. Chen Y, Liu J, Zhi S, et al. Repurposing type I-F CRISPR-Cas system as a transcriptional tivation tool in human cells[J]. Nat Commun, 2020,19;11(1):3136.
[8]. Makarova KS, Wolf YI, Iranzo J, et al. Evolutionary classification of CRISPR-Cas systems: burst of class 2 and derived variants[J]. Nat Rev Microbiol, 2020,18(2):67-83.
[9]. Kantor A, McClements ME, MacLaren RE. CRISPR-Cas9 DNA Base-Editing and Prime-diting[J]. Int J Mol Sci, 2020,28;21(17):6240.
[10]. Wang Yangkun, Ji Shixin, Su Yingying, et al. Comparison and application of gene editing tools CRISPR-Cas9 and CRISPR-Cas12a[J].Journal of Changchun Normal University, 2023, 42(12):107-112.
[11]. Hoy, S. M. Exagamglogene Autotemcel: First Approval[J]. Mol. Diagn. Ther. 28, 133–139 (2024).
[12]. Ding Yidan, Chen Chen. Research progress of CRISPR-Cas9 gene editing technology in tumor treatment[J]. Acta Laser Biologica Sinica,2022,31(06):488-497.
[13]. Suh, S., Choi, E. H., et al. Precision genome editing in the eye[J]. Proc. Natl. Acad. Sci, 2022,119, e2210104119.
[14]. Jiang J, Kong K, Fang X, et al. CRISPR-Cas9-mediated deletion of carbonic anhydrase 2 in the ciliary body to treat glaucoma[J]. Cell Rep Med, 2024,21;5(5):101524.
[15]. Fang Shuai, Wang Gang, Yang Linhua. Research progress of AAV-CRISPR/Cas9-mediated gene therapy for hemophilia A[J].Chinese Journal of Experimental Hematology,2023,31(06):1890-1893.
[16]. Bulfield G, Siller WG, Wight PA, Moore KJ. X chromosome-linked muscular dystrophy (mdx) in the mouse[J]. Proc Natl Acad Sci U S A, 1984,81(4):1189-92.
[17]. Zhu Yuling, Sun Yiming, Zhang Huili, et al. Neuromuscular junction characteristics of mice in Duchenne muscular dystrophy model[J].Chinese Journal of Modern Neurological Diseases, 2015, 15(05):374-379.
[18]. Zhang Y, Nishiyama T, Olson EN, Bassel-Duby R. CRISPR/Cas correction of muscular dystrophies[J]. Exp Cell Res, 2021,1;408(1):112844.
[19]. Wang Danni. Establishment of Duchenne muscular dystrophy comprehensive database and dystrophin gene editing based on CRISPR/Cas9 technology[D].Fujian Medical University, 2017.
[20]. Rashnonejad, A. et al. FP.29 AAV-CRISPR-Cas13 gene therapy for FSHD: DUX4 gene silencing efficacy and immune responses to Cas13b protein[J]. Neuromuscul. Disord, 2022,32, S103–S104.
Cite this article
Zhang,H. (2024). CRISPR/Cas system in human genetic diseases. Theoretical and Natural Science,50,61-67.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of ICBioMed 2024 Workshop: Workshop on Intelligent Medical Data Analysis for Precision Medicine
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Townsend JA, Wright DA, Winfrey RJ,et al. High-frequency modification of plant genes sing engineered zinc-finger nucleases[J]. Nature, 2009, 21;459(7245):442-5.
[2]. Joung JK, Sander JD. TALENs: a widely applicable technology for targeted genome editing[J]. at Rev Mol Cell Biol, 2013,14(1):49-55.
[3]. Wang Linlin, Li Hongling. Application of CRISPR/Cas9 gene editing technology in precision ncology research[J].Chinese Journal of Cancer Biotherapy, 2024,31(05):519-527.
[4]. Xue Shan, Wang Shuya, Liu Li, et al. Research progress on precision gene editing technology ased on CRISPR/Cas9[J].Chinese Journal of Biotechnology, 2023,39(07):2566-2578.
[5]. Wong, C. UK first to approve CRISPR treatment for diseases: what you need to know[J]. ature, 2023, 623, 676–677.
[6]. Xin Yuan, Tian Kairen, Qiao Jianjun, et al. Gene editing tools based on CRISPR/Cas system nd their improvement strategies[J].Chinese Journal of Biotechnology, 2023,43(08):72-85.
[7]. Chen Y, Liu J, Zhi S, et al. Repurposing type I-F CRISPR-Cas system as a transcriptional tivation tool in human cells[J]. Nat Commun, 2020,19;11(1):3136.
[8]. Makarova KS, Wolf YI, Iranzo J, et al. Evolutionary classification of CRISPR-Cas systems: burst of class 2 and derived variants[J]. Nat Rev Microbiol, 2020,18(2):67-83.
[9]. Kantor A, McClements ME, MacLaren RE. CRISPR-Cas9 DNA Base-Editing and Prime-diting[J]. Int J Mol Sci, 2020,28;21(17):6240.
[10]. Wang Yangkun, Ji Shixin, Su Yingying, et al. Comparison and application of gene editing tools CRISPR-Cas9 and CRISPR-Cas12a[J].Journal of Changchun Normal University, 2023, 42(12):107-112.
[11]. Hoy, S. M. Exagamglogene Autotemcel: First Approval[J]. Mol. Diagn. Ther. 28, 133–139 (2024).
[12]. Ding Yidan, Chen Chen. Research progress of CRISPR-Cas9 gene editing technology in tumor treatment[J]. Acta Laser Biologica Sinica,2022,31(06):488-497.
[13]. Suh, S., Choi, E. H., et al. Precision genome editing in the eye[J]. Proc. Natl. Acad. Sci, 2022,119, e2210104119.
[14]. Jiang J, Kong K, Fang X, et al. CRISPR-Cas9-mediated deletion of carbonic anhydrase 2 in the ciliary body to treat glaucoma[J]. Cell Rep Med, 2024,21;5(5):101524.
[15]. Fang Shuai, Wang Gang, Yang Linhua. Research progress of AAV-CRISPR/Cas9-mediated gene therapy for hemophilia A[J].Chinese Journal of Experimental Hematology,2023,31(06):1890-1893.
[16]. Bulfield G, Siller WG, Wight PA, Moore KJ. X chromosome-linked muscular dystrophy (mdx) in the mouse[J]. Proc Natl Acad Sci U S A, 1984,81(4):1189-92.
[17]. Zhu Yuling, Sun Yiming, Zhang Huili, et al. Neuromuscular junction characteristics of mice in Duchenne muscular dystrophy model[J].Chinese Journal of Modern Neurological Diseases, 2015, 15(05):374-379.
[18]. Zhang Y, Nishiyama T, Olson EN, Bassel-Duby R. CRISPR/Cas correction of muscular dystrophies[J]. Exp Cell Res, 2021,1;408(1):112844.
[19]. Wang Danni. Establishment of Duchenne muscular dystrophy comprehensive database and dystrophin gene editing based on CRISPR/Cas9 technology[D].Fujian Medical University, 2017.
[20]. Rashnonejad, A. et al. FP.29 AAV-CRISPR-Cas13 gene therapy for FSHD: DUX4 gene silencing efficacy and immune responses to Cas13b protein[J]. Neuromuscul. Disord, 2022,32, S103–S104.









