1. Introduction
Cancer is one of the leading causes of death worldwide today. According to statistics from the World Health Organization, there were 19.29 million new cancer cases and 9.96 million deaths worldwide in 2020[1]. Although cancer diagnosis and treatment methods have continued to improve in recent years, the prognosis of most cancer patients is still poor, and the 5-year survival rate is generally not high. Therefore, in-depth study of the molecular mechanisms of cancer occurrence and development and finding key targets for inhibiting cancer are of great significance for improving the prognosis of cancer patients.
The cell cycle is the basis of eukaryotic cell proliferation and plays a vital role in maintaining the homeostasis of body tissues. Cell cycle disorder is one of the important characteristics of cancer [2]. A large number of studies have shown that abnormal expression of cell cycle-related genes and proteins is common in various cancers, leading to imbalance of cell cycle checkpoints, causing excessive cell proliferation and causing tumors[3]. Therefore, abnormal cell cycle regulation is considered to be a key driver of cancer development. The development of anticancer drugs targeting cell cycle-related molecules has become an important direction in cancer treatment research.
DTL (denticleless E3 ubiquitin protein ligase homolog) is an important cell cycle regulator discovered in recent years. DTL belongs to the DCAF (DDB1 and CUL4 associated factors) family and can serve as a substrate receptor for CRL4 (cullin-RING ubiquitin ligase 4), mediating the ubiquitination and degradation of a variety of cell cycle-related proteins, such as CDT1, p21, Set8, etc. , thus playing an important role in DNA replication, DNA damage repair, cell cycle checkpoints and other processes [4,5]. Studies have found that DTL is highly expressed in a variety of human tumor tissues, suggesting that it may play an important role in the occurrence and development of cancer. Studies have confirmed that DTL inhibits DNA re-replication by regulating the degradation of CDT1, maintains genome stability, and plays a key role in inhibiting tumorigenesis [6]. In addition, DTL can also degrade p21 and Set8 through ubiquitination, relieve G1/S and G2/M checkpoint blocks, and promote cell cycle progression [7,8]. These functional abnormalities of DTL may be its important pathogenic mechanism in cancer.
In view of the important role of DTL in cell cycle regulation and cancer occurrence, in-depth elucidation of the molecular mechanism of DTL and its relationship with cancer is of great significance for the prevention, diagnosis and treatment of cancer. This article will systematically summarize the research progress of DTL, focus on the role and mechanism of DTL in cell cycle regulation and cancer development, and look forward to its application prospects as a new target for anti-cancer drugs, providing a reference for future DTL-related cancer research.
2. DTL overexpression promotes cancer progression
DTL (denticleless E3 ubiquitin protein ligase homolog) is an E3 ubiquitin ligase that is highly expressed in a variety of tumors and is closely related to the occurrence and development of tumors. Studies have shown that miR-203 can target the 3’UTR of DTL. region, inhibiting its expression [9]. The overexpression of DTL can degrade multiple tumor suppressor genes through ubiquitination, thereby promoting the proliferation, invasion and metastasis of tumor cells.
PDCD4 (programmed cell death 4) is an important tumor suppressor gene that can inhibit the proliferation and invasion of tumor cells [10,11]. Studies have found that DTL can directly interact with PDCD4 and mediate its ubiquitination and degradation [12]. Overexpression of DTL leads to a significant decrease in PDCD4 protein levels, thereby releasing the inhibitory effect of PDCD4 on tumor cell proliferation and invasion and promoting tumor progression. In addition, DTL can also induce epithelial epithelial cell death by activating the RAC1-JNK-FOXO1 signaling pathway. Mesenchymal transition (EMT), enhances the migration and invasion ability of tumor cells [13]. In addition to PDCD4, DTL can also ubiquitinate and degrade another tumor suppressor gene p21 [14]. p21 is a cyclin-dependent kinase inhibitor. It can block the cell cycle process. By degrading p21, DTL relieves its inhibition on the cell cycle and promotes tumor cell proliferation [14]. In addition, DTL can also induce epithelial-mesenchymal transition by activating the RAC1-JNK-FOXO1 signaling pathway ( EMT), enhancing the migration and invasion capabilities of tumor cells [13].
In recent years, more and more studies have shown that the DTL (Denticleless E3 Ubiquitin Protein Ligase) gene is overexpressed in various cancers such as lung cancer, liver cancer, and gastric cancer, and promotes cancer progression.
In 2008, Ueki et al. [15] first reported that DTL is highly expressed in breast cancer tissues, and knocking down DTL can inhibit the proliferation and invasion of breast cancer cells. Subsequently, Perez-Peaamp et al. [16] used gene expression and functional annotation analysis to identify genes differentially expressed in the ubiquitin pathway between normal breast and basal-like tumors and found that UBE2T and DTL were expanded in approximately 12% of breast tumors. increase.
Hepatocellular carcinoma is another common tumor with overexpression of DTL. Li et al. found that DTL is highly expressed in liver cancer tissues and is related to tumor stage, vascular invasion and recurrence. The overall survival of liver cancer patients with high expression of DTL is significantly shortened. In terms of clinical manifestations, DTL is related to the overall survival of HCC patients, and patients with high DTL expression have shorter survival times. Mechanistic studies have shown that DTL deletion leads to the destruction of mitotic proteins and the upregulation of the cell cycle arrest gene p21, and targeting DTL reduces cell cycle regulatory factors and chromosome segregation genes, resulting in an increase in cell micronuclei. In this regard, Chen et al. suggested inhibiting the growth of cancer cells through down-regulation of TPX2 [17]. The mRNA expression of the CUL4 complex, including DTL, is increased, and patient survival time is poor. The final experiment showed that E2F1 mediates DTL to promote the invasion and metastasis of liver cancer cells [18], which also provides evidence for DTL as a prognostic marker and therapeutic target for liver cancer. points provide the basis.
In non-small cell lung cancer, miR-203 is transferred into tumor cells through extracellular vesicles (EVs) secreted by human umbilical vein endothelial cells (HUVECs), targeting the expression of DTL, thereby upregulating the protein level of PDCD4, inhibiting Malignant phenotype of tumor cells[9,10,19].
Regarding the role of DTL in colorectal cancer, Baraniskin et al. found in 2014 [20] that DTL mRNA is highly expressed in colorectal cancer tissue and is positively correlated with tumor stage. Knocking down DTL inhibits the proliferation and colony formation ability of colorectal cancer cells. In 2017, Kobayashi et al. [21] reported that DTL promotes epithelial-to-mesenchymal transition (EMT) and metastasis of colorectal cancer cells by regulating the Wnt/β-catenin pathway.
In cervical adenocarcinoma, high expression of DTL is associated with depth of tumor invasion, lymph node metastasis, and poor prognosis [13]. These findings suggest that DTL may become a potential therapeutic target against metastasis of cervical adenocarcinoma [11]. 2023 Luo et al. [22] used real-time quantitative polymerase chain reaction (qRT-PCR) to detect the expression of circ-acyclic E3 ubiquitin protein ligase homolog (circ-DTL), miR-758-3p and DCUN1D1. It was found that Circ-DTL acts as a tumor promoter in cervical cancer development by regulating the miR-758-3p/DCUN1D1 pathway. Knockdown of circ-DTL can inhibit the growth, migration and invasion of cervical cancer cells, and promote cell cycle arrest and apoptosis. In bladder cancer, Luo confirmed through in vitro and in vivo experiments that DTL may promote BCa progression through the AKT/mTOR pathway[23].
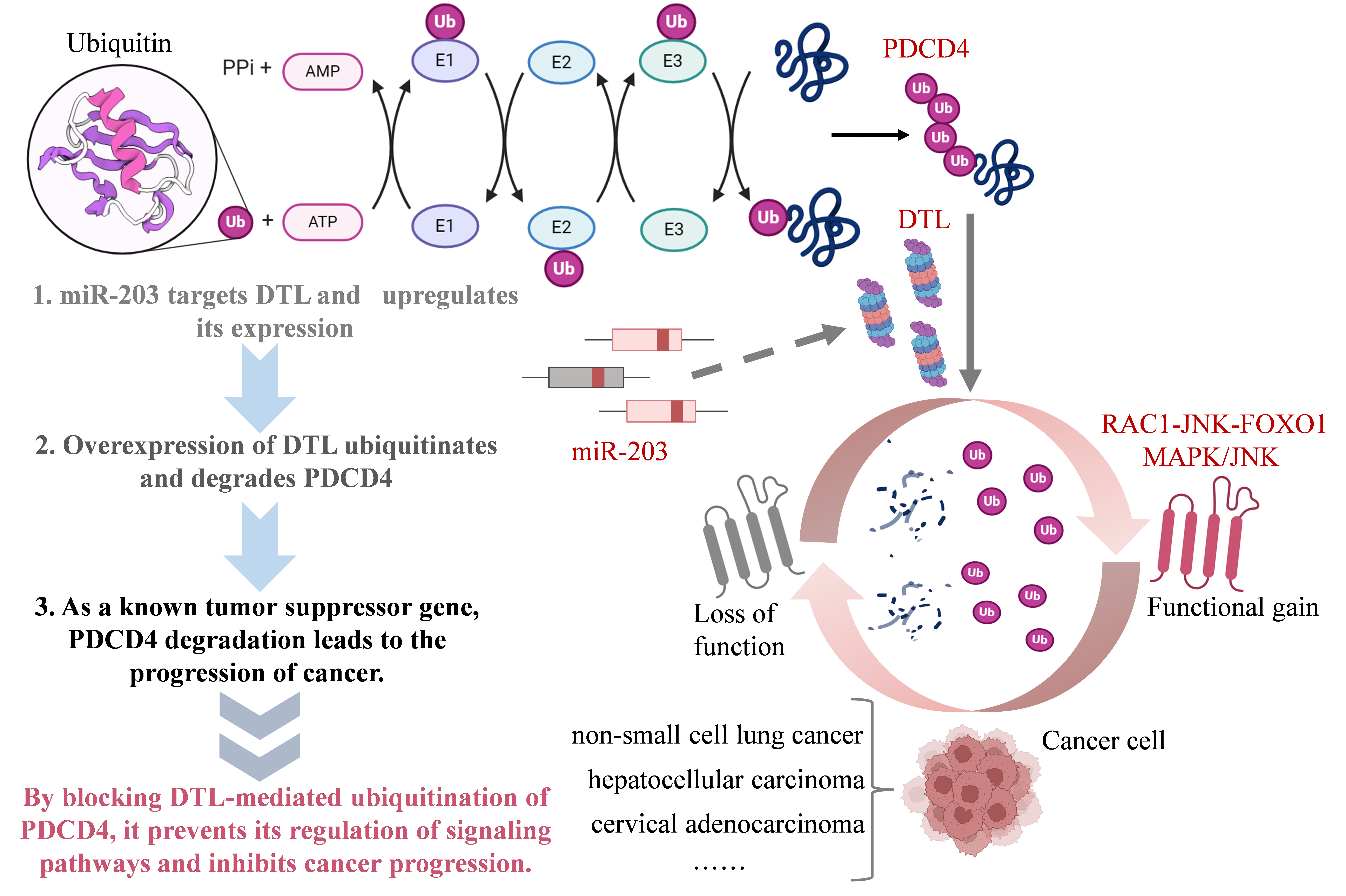
Figure 1. Mechanisms of pro-cancer response by miR-203→DTL→PDCD4 axis
The above views all confirm that the “miR-203→DTL→PDCD4” reaction chain plays an important role in tumor progression (Figure 3.1). miR-203 targets the expression of DTL, and DTL overexpression can ubiquitinate and degrade multiple Tumor suppressor genes such as PDCD4, p21, etc. relieve their inhibitory effects on tumor cell proliferation, invasion and metastasis, and promote tumor progression through various mechanisms such as inducing EMT and reshaping the tumor immune microenvironment[10,13,19]. In-depth Studying the miRNA-DTL-tumor suppressor gene axis will help elucidate the molecular mechanism of tumor development and provide new ideas and strategies for tumor diagnosis, prognosis assessment and targeted therapy. In addition to the above-mentioned cancers, DTL is also involved in melanoma [24], colorectal cancer [20], ovarian cancer[16] and other cancers are also highly expressed, promoting the proliferation, invasion, migration and drug resistance of cancer cells. In the future, more clinical samples and animal model experiments are needed to further verify the feasibility and effectiveness of DTL as a tumor marker and therapeutic target.
3. The regulatory mechanism of DTL on the cell cycle
DTL is also a protein that plays an important role in cell cycle regulation. The latest research shows that DTL mainly maintains genome stability through two different mechanisms. On the one hand, DTL is an important component of the CUL4-DDB1 ubiquitin ligase complex, to prevent repeated DNA replication by regulating the level of CDT1 [6]; on the other hand, DTL is also essential in the G2/M checkpoint activation process induced by DNA damage [6].
During normal cell growth, DTL forms a complex with CUL4-DDB1 and degrades CDT1 through the ubiquitination pathway, thus inhibiting the initiation of DNA replication mediated by CDT1 and preventing repeated replication of the genome [6,25]. CDT1 is a DNA replication It is necessary for the formation of the pre-replication complex, and the strict regulation of CDT1 levels is crucial to ensure that DNA is only replicated once per cell cycle. Studies have found that knocking down DTL will lead to an increase in CDT1 protein levels, causing Part of the DNA is replicated repeatedly, eventually leading to an increase in DNA content >4N [6]. In addition, DTL deletion can also lead to a series of phenotypes such as delayed G2 phase, excessive centromere replication, and spindle multipolarization. These phenotypes It is very similar to the overexpression of CDT1 or the deletion of Geminin (the inhibitor of CDT1)[26,27]. These results strongly prove that the DTL-CUL4-DDB1 complex inhibits DNA repeat replication and maintains genome stability by degrading CDT1.
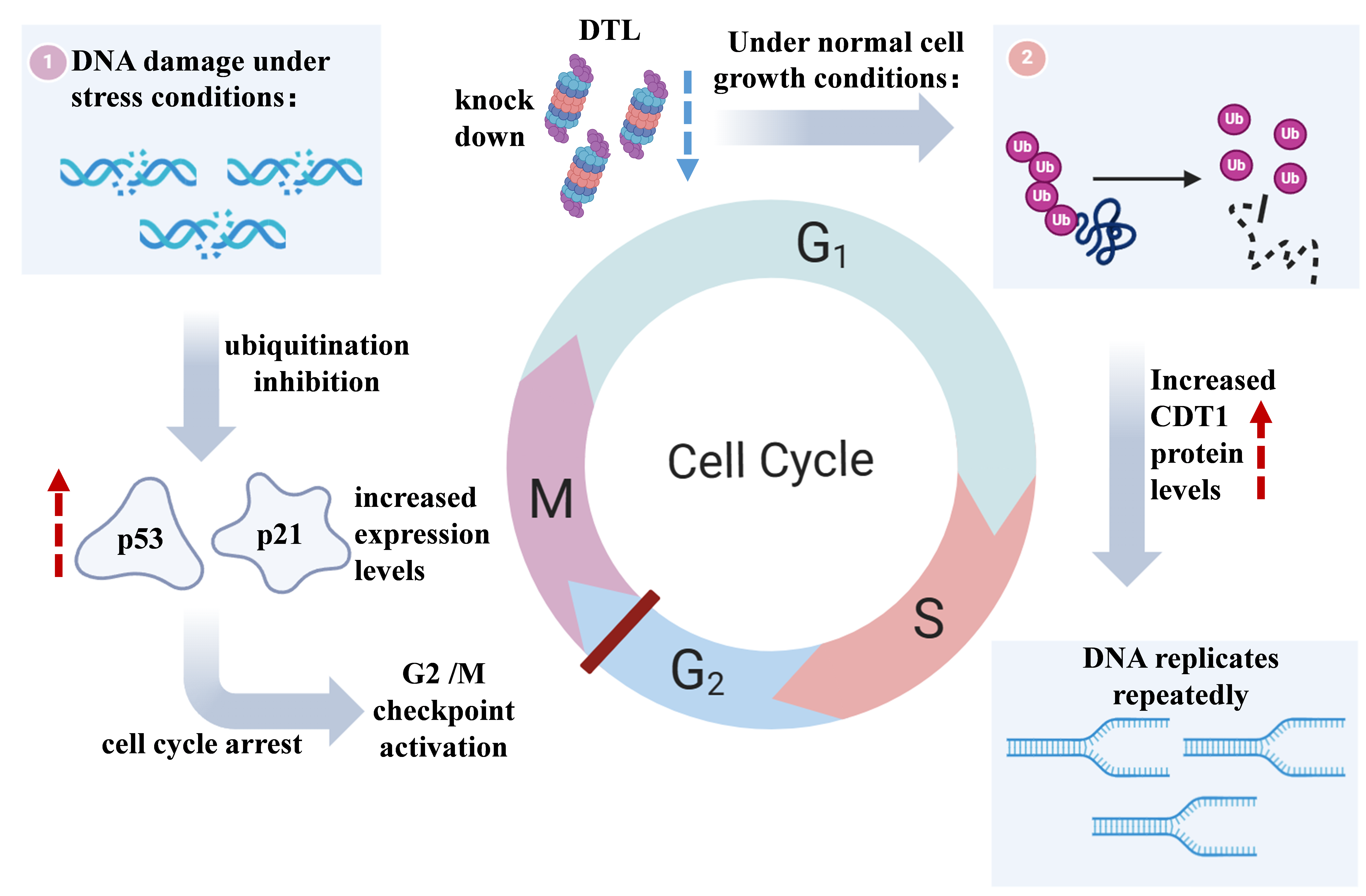
Figure 2. Two main mechanisms by which DTL regulates the cell cycle
In addition to regulating CDT1, DTL also plays an irreplaceable role in the G2/M checkpoint induced by DNA damage. Studies have found in zebrafish embryos that DTL mutants cannot effectively block the mitotic process after ionizing radiation treatment, showing showed obvious G2/M checkpoint defects [6]. Further knocking down DTL in human cells also destroyed the G2/M checkpoint induced by ionizing radiation. Interestingly, this function does not seem to be related to CDT1, because in Although knocking down DTL and CDT1 simultaneously in zebrafish embryos can rescue the cell cycle defects caused by DTL deletion, it cannot restore the G2/M checkpoint. This suggests that DTL may activate the DNA damage checkpoint by regulating other substrates[25]. The latest research shows that DTL may trigger G2 arrest by stabilizing p53 and p21 [25]. DTL interacts with the MDM2-p53 ubiquitination complex, and knocking down DTL will lead to the inhibition of p53 ubiquitination, p53 and p21 levels increase, thereby causing cell cycle arrest [26].
4. Summary
In summary, as an important component of the CUL4-DDB1 ubiquitin ligase complex, DTL mainly maintains genome stability in two ways: first, under DNA damage stress conditions, DTL participates in the G2/M checkpoint Activation blocks the mitosis process and buys time for DNA repair; secondly, during normal cell growth, DTL participates in ubiquitination and degradation of CDT1 to prevent repeated DNA replication. These two processes complement each other and jointly ensure the integrity and stability of the genome. The loss of DTL function will lead to increased genome instability and may promote the occurrence and development of tumors. Therefore, it is necessary to conduct in-depth research on the molecular mechanism of DTL and its relationship with tumorigenesis. , is of great significance for the prevention, diagnosis and treatment of tumors. Future work also needs to further elucidate the regulatory network between DTL and other cell cycle and DNA repair factors, as well as the role and mechanism of DTL abnormalities in tumorigenesis.
With the rapid development of single-cell level methods at this stage, whole-genome visualization and imaging technology enables the intuitive characterization of gene expression and the spatial organization and folding of the genome in various types of cells. Single-cell transcriptomics (scRNA-seq) [28–30] and single-cell chromosome conformation capture technology (scHi-C) [31,32] provide a new perspective and powerful tool for in-depth understanding of the role of DTL in the development of cancer. The tumor microenvironment plays a key role in tumor progression [33–35]. Using scRNA-seq technology to analyze the expression pattern of DTL in tumor cells and microenvironment cells (such as immune cells, stromal cells) will help understand the interaction between DTL expression and the tumor microenvironment, and provide clues for the development of new cancer immunotherapy strategies[36,37]. Faced with the heterogeneity caused by multicellular subtypes in tumor tissue, using scRNA-seq technology to analyze the expression patterns of DTL in cells of different tumor subtypes will help to understand its role in the formation and maintenance of tumor heterogeneity. Combining scRNA-seq and scHi-C for DTL regulatory network analysis, scRNA-seq can provide gene expression information at the single cell level, while scHi-C can reveal the relationship between the 3D structure of chromosomes and gene expression regulation. By integrating the two types of data [38], a multi-modal regulatory network of DTL at the single cell level can be constructed [39], which opens up broad prospects for exploring DTL as a cancer marker and therapeutic target. Future research should make full use of these new technologies to understand the role of DTL in tumor occurrence and development in a multi-dimensional and dynamic manner, build a more accurate prognostic model, and explore more effective prevention and treatment strategies, ultimately benefiting cancer patients.
References
[1]. Sung H, Ferlay J, Siegel R L, et al. Global cancer statistics 2020: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA: A Cancer Journal for Clinicians, 2021, 71(3): 209–249. DOI:10.3322/caac.21660.
[2]. Hanahan D, Weinberg R A. Hallmarks of cancer: the next generation[J]. Cell, 2011, 144(5): 646–674. DOI:10.1016/j.cell.2011.02.013.
[3]. Otto T, Sicinski P. Cell cycle proteins as promising targets in cancer therapy[J]. Nature Reviews Cancer, 2017, 17(2): 93–115. DOI:10.1038/nrc.2016.138.
[4]. Abbas T, Dutta A. CRL4 Cdt2: master coordinator of cell cycle progression and genome stability[J]. Cell Cycle, 2011, 10(2): 241–249. DOI:10.4161/cc.10.2.14530.
[5]. Havens C G, Walter J C. Mechanism of crl4 Cdt2 , a pcna-dependent e3 ubiquitin ligase[J]. Genes & Development, 2011, 25(15): 1568–1582. DOI:10.1101/gad.2068611.
[6]. Sansam C L, Shepard J L, Lai K, et al. DTL/cdt2 is essential for both cdt1 regulation and the early g2/m checkpoint[J]. Genes & Development, 2006, 20(22): 3117–3129. DOI:10.1101/gad. 1482106.
[7]. Kim Y, Starostina N G, Kipreos E T. The crl4 Cdt2 ubiquitin ligase targets the degradation of p21 Cip1 to control replication licensing[J]. Genes & Development, 2008, 22(18): 2507–2519. DOI:10.1101/gad.1703708.
[8]. Abbas T, Shibata E, Park J, et al. CRL4Cdt2 regulates cell proliferation and histone gene expression by targeting pr-set7/set8 for degradation[J]. Molecular Cell, 2010, 40(1): 9–21. DOI:10.1016/j.molcel.2010.09.014.
[9]. Ma T, Hu Y, Guo Y, et al. Human umbilical vein endothelial cells-derived microrna-203-containing extracellular vesicles alleviate non-small-cell lung cancer progression through modulating the dtl/p21 axis[J]. Cancer Gene Therapy, 2022, 29(1): 87–100. DOI:10.1038/s41417-020-00292-3.
[10]. Fan Q, Lu Q, Wang G, et al. Optimizing component formula suppresses lung cancer by blocking dtl-mediated pdcd4 ubiquitination to regulate the mapk/jnk pathway[J]. Journal of Ethnopharmacology, 2022, 299: 115546. DOI:10.1016/j.jep.2022.115546.
[11]. Zhang X, Wang X, Song X, et al. Programmed cell death 4 enhances chemosensitivity of ovarian cancer cells by activating death receptor pathway in vitro and in vivo[J]. Cancer Science, 2010, 101(10): 2163–2170. DOI:10.1111/j.1349-7006.2010.01664.x.
[12]. Cui H, Wang Q, Lei Z, et al. DTL promotes cancer progression by pdcd4 ubiquitin-dependent degradation[J]. Journal of Experimental & Clinical Cancer Research, 2019, 38(1): 350. DOI:10.1186/s13046-019-1358-x.
[13]. Liu S, Gu L, Wu N, et al. Overexpression of dtl enhances cell motility and promotes tumor metastasis in cervical adenocarcinoma by inducing rac1-jnk-foxo1 axis[J]. Cell Death & Disease, 2021, 12(10): 929. DOI:10.1038/s41419-021-04179-5.
[14]. Abbas T, Dutta A. P21 in cancer: intricate networks and multiple activities[J]. Nature Reviews Cancer, 2009, 9(6): 400–414. DOI:10.1038/nrc2657.
[15]. Ueki T, Nishidate T, Park J H, et al. Involvement of elevated expression of multiple cell-cycle regulator, dtl/ramp (denticleless/ra-regulated nuclear matrix associated protein), in the growth of breast cancer cells[J]. Oncogene, 2008, 27(43): 5672–5683. DOI:10.1038/onc.2008.186.
[16]. Perez-Peamp J. Ubiquitin-conjugating enzyme e2t (ube2t) and denticleless protein homolog (dtl) are linked to poor outcome in breast and lung cancers[J]. SCIENTIfIC REpOrts, [no date].
[17]. Chen Y-C, Chen I, Huang G-J, et al. Targeting dtl induces cell cycle arrest and senescence and suppresses cell growth and colony formation through tpx2 inhibition in human hepatocellular carcinoma cells[J]. OncoTargets and Therapy, 2018, Volume 11: 1601–1616. DOI:10.2147/OTT.S147453.
[18]. Dong R, Zhang D, Han B, et al. DTL is a novel downstream gene of e2f1 that promotes the progressionof hepatocellular carcinoma[J]. Current Cancer Drug Targets, 2023, 23(10): 817–828. DOI:10.2174/1568009623666230511100246.
[19]. Pan X, Chen S, Ye L, et al. Long non-coding rna dlgap1-as1 modulates the development of non-small-cell lung cancer via the microrna-193a-5p/dtl axis[J]. Laboratory Investigation, 2022, 102(11): 1182–1191. DOI:10.1038/s41374-022-00831-6.
[20]. Baraniskin A, Birkenkamp-Demtroder K, Maghnouj A, et al. MiR-30a-5p suppresses tumor growth in colon carcinoma by targeting dtl[J]. Carcinogenesis, 2012, 33(4): 732–739. DOI:10.1093/carcin/bgs020.
[21]. Kobayashi H, Komatsu S, Ichikawa D, et al. Overexpression of denticleless e3 ubiquitin protein ligase homolog (dtl) is related to poor outcome in gastric carcinoma[J]. Oncotarget, 2015, 6(34): 36615–36624. DOI:10.18632/oncotarget.5620.
[22]. Luo X, Liu J, Wang X, et al. Circ‐DTL sponges mir‐758‐3p to accelerate cervical cancer malignant progression by regulating dcun1d1 expression[J]. Journal of Biochemical and Molecular Toxicology, 2023, 37(11): e23462. DOI:10.1002/jbt.23462.
[23]. Luo Y, He Z, Liu W, et al. DTL is a prognostic biomarker and promotes bladder cancer progression through regulating the akt/mtor axis[J]. Oxidative Medicine and Cellular Longevity, [no date].
[24]. Lu J-J, Chen F-J, Li Y, et al. DTL promotes melanoma progression through rewiring cell glucose metabolism[J]. Annals of Translational Medicine, 2022, 10(2): 68–68. DOI:10.21037/atm-21-6648.
[25]. Higa L A, Banks D, Wu M, et al. L2DTL/cdt2 interacts with the cul4/ddb1 complex and pcna and regulates cdt1 proteolysis in response to dna damage[J]. Cell Cycle, 2006, 5(15): 1675–1680. DOI:10.4161/cc.5.15.3149.
[26]. Melixetian M, Ballabeni A, Masiero L, et al. Loss of geminin induces rereplication in the presence of functional p53[J]. The Journal of Cell Biology, 2004, 165(4): 473–482. DOI:10.1083/jcb.200403106.
[27]. Tachibana K K, Gonzalez M A, Guarguaglini G, et al. Depletion of licensing inhibitor geminin causes centrosome overduplication and mitotic defects[J]. EMBO Reports, 2005, 6(11): 1052–1057. DOI:10.1038/sj.embor.7400527.
[28]. Gupta I, Collier P G, Haase B, et al. Single-cell isoform rna sequencing characterizes isoforms in thousands of cerebellar cells[J]. Nature Biotechnology, 2018, 36(12): 1197–1202. DOI:10.1038/nbt.4259.
[29]. Svensson V, Vento-Tormo R, Teichmann S A. Exponential scaling of single-cell rna-seq in the past decade[J]. Nature Protocols, 2018, 13(4): 599–604. DOI:10.1038/nprot.2017.149.
[30]. Mereu E, Lafzi A, Moutinho C, et al. Benchmarking single-cell rna-sequencing protocols for cell atlas projects[J]. Nature Biotechnology, 2020, 38(6): 747–755. DOI:10.1038/s41587-020-0469-4.
[31]. Ulianov S V, Razin S V. The two waves in single-cell 3d genomics[J]. Seminars in Cell & Developmental Biology, 2022, 121: 143–152. DOI:10.1016/j.semcdb.2021.05.021.
[32]. Zhou T, Zhang R, Ma J. The 3d genome structure of single cells[J]. Annual Review of Biomedical Data Science, 2021, 4(1): 21–41. DOI:10.1146/annurev-biodatasci-020121-084709.
[33]. Chen K, Wang Q, Li M, et al. Single-cell rna-seq reveals dynamic change in tumor microenvironment during pancreatic ductal adenocarcinoma malignant progression[J]. EBioMedicine, 2021, 66: 103315. DOI:10.1016/j.ebiom.2021.103315.
[34]. Raghavan S, Winter P S, Navia A W, et al. Microenvironment drives cell state, plasticity, and drug response in pancreatic cancer[J]. Cell, 2021, 184(25): 6119-6137.e26. DOI:10.1016/j.cell. 2021.11.017.
[35]. Tikhonova A N, Dolgalev I, Hu H, et al. The bone marrow microenvironment at single-cell resolution[J]. Nature, 2019, 569(7755): 222–228. DOI:10.1038/s41586-019-1104-8.
[36]. Perez-Peña J, Corrales-Sánchez V, Amir E, et al. Ubiquitin-conjugating enzyme e2t (ube2t) and denticleless protein homolog (dtl) are linked to poor outcome in breast and lung cancers[J]. Scientific Reports, 2017, 7(1): 17530. DOI:10.1038/s41598-017-17836-7.
[37]. Werba G, Weissinger D, Kawaler E A, et al. Single-cell rna sequencing reveals the effects of chemotherapy on human pancreatic adenocarcinoma and its tumor microenvironment[J]. Nature Communications, 2023, 14(1): 797. DOI:10.1038/s41467-023-36296-4.
[38]. Liu Z, Chen Y, Xia Q, et al. Linking genome structures to functions by simultaneous single-cell hi-c and rna-seq: 6649[J]. Science, 2023, 380(6649): 1070–1076. DOI:10.1126/science. adg3797.
[39]. Yousuf S, Qiu M, Voith Von Voithenberg L, et al. Spatially resolved multi-omics single-cell analyses inform mechanisms of immune dysfunction in pancreatic cancer[J]. Gastroenterology, 2023, 165(4): 891-908.e14. DOI:10.1053/j.gastro.2023.05.036.
Cite this article
Yue,S.;Fang,Y. (2024). Research on the mechanism of DTL in the occurrence and development of cancer. Theoretical and Natural Science,49,53-59.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 4th International Conference on Biological Engineering and Medical Science
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Sung H, Ferlay J, Siegel R L, et al. Global cancer statistics 2020: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries[J]. CA: A Cancer Journal for Clinicians, 2021, 71(3): 209–249. DOI:10.3322/caac.21660.
[2]. Hanahan D, Weinberg R A. Hallmarks of cancer: the next generation[J]. Cell, 2011, 144(5): 646–674. DOI:10.1016/j.cell.2011.02.013.
[3]. Otto T, Sicinski P. Cell cycle proteins as promising targets in cancer therapy[J]. Nature Reviews Cancer, 2017, 17(2): 93–115. DOI:10.1038/nrc.2016.138.
[4]. Abbas T, Dutta A. CRL4 Cdt2: master coordinator of cell cycle progression and genome stability[J]. Cell Cycle, 2011, 10(2): 241–249. DOI:10.4161/cc.10.2.14530.
[5]. Havens C G, Walter J C. Mechanism of crl4 Cdt2 , a pcna-dependent e3 ubiquitin ligase[J]. Genes & Development, 2011, 25(15): 1568–1582. DOI:10.1101/gad.2068611.
[6]. Sansam C L, Shepard J L, Lai K, et al. DTL/cdt2 is essential for both cdt1 regulation and the early g2/m checkpoint[J]. Genes & Development, 2006, 20(22): 3117–3129. DOI:10.1101/gad. 1482106.
[7]. Kim Y, Starostina N G, Kipreos E T. The crl4 Cdt2 ubiquitin ligase targets the degradation of p21 Cip1 to control replication licensing[J]. Genes & Development, 2008, 22(18): 2507–2519. DOI:10.1101/gad.1703708.
[8]. Abbas T, Shibata E, Park J, et al. CRL4Cdt2 regulates cell proliferation and histone gene expression by targeting pr-set7/set8 for degradation[J]. Molecular Cell, 2010, 40(1): 9–21. DOI:10.1016/j.molcel.2010.09.014.
[9]. Ma T, Hu Y, Guo Y, et al. Human umbilical vein endothelial cells-derived microrna-203-containing extracellular vesicles alleviate non-small-cell lung cancer progression through modulating the dtl/p21 axis[J]. Cancer Gene Therapy, 2022, 29(1): 87–100. DOI:10.1038/s41417-020-00292-3.
[10]. Fan Q, Lu Q, Wang G, et al. Optimizing component formula suppresses lung cancer by blocking dtl-mediated pdcd4 ubiquitination to regulate the mapk/jnk pathway[J]. Journal of Ethnopharmacology, 2022, 299: 115546. DOI:10.1016/j.jep.2022.115546.
[11]. Zhang X, Wang X, Song X, et al. Programmed cell death 4 enhances chemosensitivity of ovarian cancer cells by activating death receptor pathway in vitro and in vivo[J]. Cancer Science, 2010, 101(10): 2163–2170. DOI:10.1111/j.1349-7006.2010.01664.x.
[12]. Cui H, Wang Q, Lei Z, et al. DTL promotes cancer progression by pdcd4 ubiquitin-dependent degradation[J]. Journal of Experimental & Clinical Cancer Research, 2019, 38(1): 350. DOI:10.1186/s13046-019-1358-x.
[13]. Liu S, Gu L, Wu N, et al. Overexpression of dtl enhances cell motility and promotes tumor metastasis in cervical adenocarcinoma by inducing rac1-jnk-foxo1 axis[J]. Cell Death & Disease, 2021, 12(10): 929. DOI:10.1038/s41419-021-04179-5.
[14]. Abbas T, Dutta A. P21 in cancer: intricate networks and multiple activities[J]. Nature Reviews Cancer, 2009, 9(6): 400–414. DOI:10.1038/nrc2657.
[15]. Ueki T, Nishidate T, Park J H, et al. Involvement of elevated expression of multiple cell-cycle regulator, dtl/ramp (denticleless/ra-regulated nuclear matrix associated protein), in the growth of breast cancer cells[J]. Oncogene, 2008, 27(43): 5672–5683. DOI:10.1038/onc.2008.186.
[16]. Perez-Peamp J. Ubiquitin-conjugating enzyme e2t (ube2t) and denticleless protein homolog (dtl) are linked to poor outcome in breast and lung cancers[J]. SCIENTIfIC REpOrts, [no date].
[17]. Chen Y-C, Chen I, Huang G-J, et al. Targeting dtl induces cell cycle arrest and senescence and suppresses cell growth and colony formation through tpx2 inhibition in human hepatocellular carcinoma cells[J]. OncoTargets and Therapy, 2018, Volume 11: 1601–1616. DOI:10.2147/OTT.S147453.
[18]. Dong R, Zhang D, Han B, et al. DTL is a novel downstream gene of e2f1 that promotes the progressionof hepatocellular carcinoma[J]. Current Cancer Drug Targets, 2023, 23(10): 817–828. DOI:10.2174/1568009623666230511100246.
[19]. Pan X, Chen S, Ye L, et al. Long non-coding rna dlgap1-as1 modulates the development of non-small-cell lung cancer via the microrna-193a-5p/dtl axis[J]. Laboratory Investigation, 2022, 102(11): 1182–1191. DOI:10.1038/s41374-022-00831-6.
[20]. Baraniskin A, Birkenkamp-Demtroder K, Maghnouj A, et al. MiR-30a-5p suppresses tumor growth in colon carcinoma by targeting dtl[J]. Carcinogenesis, 2012, 33(4): 732–739. DOI:10.1093/carcin/bgs020.
[21]. Kobayashi H, Komatsu S, Ichikawa D, et al. Overexpression of denticleless e3 ubiquitin protein ligase homolog (dtl) is related to poor outcome in gastric carcinoma[J]. Oncotarget, 2015, 6(34): 36615–36624. DOI:10.18632/oncotarget.5620.
[22]. Luo X, Liu J, Wang X, et al. Circ‐DTL sponges mir‐758‐3p to accelerate cervical cancer malignant progression by regulating dcun1d1 expression[J]. Journal of Biochemical and Molecular Toxicology, 2023, 37(11): e23462. DOI:10.1002/jbt.23462.
[23]. Luo Y, He Z, Liu W, et al. DTL is a prognostic biomarker and promotes bladder cancer progression through regulating the akt/mtor axis[J]. Oxidative Medicine and Cellular Longevity, [no date].
[24]. Lu J-J, Chen F-J, Li Y, et al. DTL promotes melanoma progression through rewiring cell glucose metabolism[J]. Annals of Translational Medicine, 2022, 10(2): 68–68. DOI:10.21037/atm-21-6648.
[25]. Higa L A, Banks D, Wu M, et al. L2DTL/cdt2 interacts with the cul4/ddb1 complex and pcna and regulates cdt1 proteolysis in response to dna damage[J]. Cell Cycle, 2006, 5(15): 1675–1680. DOI:10.4161/cc.5.15.3149.
[26]. Melixetian M, Ballabeni A, Masiero L, et al. Loss of geminin induces rereplication in the presence of functional p53[J]. The Journal of Cell Biology, 2004, 165(4): 473–482. DOI:10.1083/jcb.200403106.
[27]. Tachibana K K, Gonzalez M A, Guarguaglini G, et al. Depletion of licensing inhibitor geminin causes centrosome overduplication and mitotic defects[J]. EMBO Reports, 2005, 6(11): 1052–1057. DOI:10.1038/sj.embor.7400527.
[28]. Gupta I, Collier P G, Haase B, et al. Single-cell isoform rna sequencing characterizes isoforms in thousands of cerebellar cells[J]. Nature Biotechnology, 2018, 36(12): 1197–1202. DOI:10.1038/nbt.4259.
[29]. Svensson V, Vento-Tormo R, Teichmann S A. Exponential scaling of single-cell rna-seq in the past decade[J]. Nature Protocols, 2018, 13(4): 599–604. DOI:10.1038/nprot.2017.149.
[30]. Mereu E, Lafzi A, Moutinho C, et al. Benchmarking single-cell rna-sequencing protocols for cell atlas projects[J]. Nature Biotechnology, 2020, 38(6): 747–755. DOI:10.1038/s41587-020-0469-4.
[31]. Ulianov S V, Razin S V. The two waves in single-cell 3d genomics[J]. Seminars in Cell & Developmental Biology, 2022, 121: 143–152. DOI:10.1016/j.semcdb.2021.05.021.
[32]. Zhou T, Zhang R, Ma J. The 3d genome structure of single cells[J]. Annual Review of Biomedical Data Science, 2021, 4(1): 21–41. DOI:10.1146/annurev-biodatasci-020121-084709.
[33]. Chen K, Wang Q, Li M, et al. Single-cell rna-seq reveals dynamic change in tumor microenvironment during pancreatic ductal adenocarcinoma malignant progression[J]. EBioMedicine, 2021, 66: 103315. DOI:10.1016/j.ebiom.2021.103315.
[34]. Raghavan S, Winter P S, Navia A W, et al. Microenvironment drives cell state, plasticity, and drug response in pancreatic cancer[J]. Cell, 2021, 184(25): 6119-6137.e26. DOI:10.1016/j.cell. 2021.11.017.
[35]. Tikhonova A N, Dolgalev I, Hu H, et al. The bone marrow microenvironment at single-cell resolution[J]. Nature, 2019, 569(7755): 222–228. DOI:10.1038/s41586-019-1104-8.
[36]. Perez-Peña J, Corrales-Sánchez V, Amir E, et al. Ubiquitin-conjugating enzyme e2t (ube2t) and denticleless protein homolog (dtl) are linked to poor outcome in breast and lung cancers[J]. Scientific Reports, 2017, 7(1): 17530. DOI:10.1038/s41598-017-17836-7.
[37]. Werba G, Weissinger D, Kawaler E A, et al. Single-cell rna sequencing reveals the effects of chemotherapy on human pancreatic adenocarcinoma and its tumor microenvironment[J]. Nature Communications, 2023, 14(1): 797. DOI:10.1038/s41467-023-36296-4.
[38]. Liu Z, Chen Y, Xia Q, et al. Linking genome structures to functions by simultaneous single-cell hi-c and rna-seq: 6649[J]. Science, 2023, 380(6649): 1070–1076. DOI:10.1126/science. adg3797.
[39]. Yousuf S, Qiu M, Voith Von Voithenberg L, et al. Spatially resolved multi-omics single-cell analyses inform mechanisms of immune dysfunction in pancreatic cancer[J]. Gastroenterology, 2023, 165(4): 891-908.e14. DOI:10.1053/j.gastro.2023.05.036.









