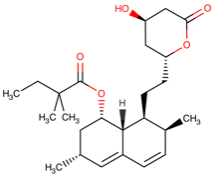
1. Introduction
Due to changes in diet, lifestyle, and a gradually aging population, cancer became more common throughout the world [1]Aging, alcoholism, hormone therapy, obesity, and genetic inheritance constituted the top risk factors for cancers [2]. Breast cancer, especially, is the most frequently diagnosed cancer in women and ranked second among cancer-related causes of death in women. It affected approximately one in eight women in high-income countries by the age of 85 years [3,4]Although many studies proved that breast cancer is treatable, more than 2 million females today globally are diagnosed with breast cancer, with nearly 700,000 death cases [5]. Current treatments for breast cancer included chemotherapy, hormone therapy, immunotherapy, radiotherapy, and surgery. When undergoing these therapies, drugs were often used as adjuvant therapies. For example, modern adjuvant chemotherapy, where drugs were discovered and combined with chemotherapy, was considered effective in treating cases of thousands of patients with hormone unresponsiveness or other complications [6]. Therefore, to explore the maximal feasibility of adjuvant therapies with drugs, drug repurposing could be used as an effective approach.
Drug repurposing is a strategy that reused approved or well-established clinical drugs for other diseases. It had been widely and successfully used in drug discovery [7]. Compared with the traditional drug development process, drug repurposing significantly improved the chance of success and reduced the cost for drug discovery in medical research. For instance, by using the drug repurposing method in the treatment of colorectal cancer, compound 19 inhibited STAT3 binding toward the hTERT promoter, indirectly inhibiting telomerase activity, and presented a specific arrest in the colorectal cancer cell cycle [8,9]. The drug repurposing process comprised four stages: the collection of candidate drugs and targets, feature representation of molecules, computational modeling and docking for the molecules, and real-world application [10].
Generating suitable drug repurposing candidates and targets was a pivotal step among the drug repurposing pipelines. As drug repurposing provided the foundational method of repositioning the drugs, recent advances in genomics, transcriptomics, proteomics, and metabolomics provided vast and deep knowledge about the molecular and metabolic alterations that occurred in cancers [11]. Specifically, single-cell RNA sequencing (scRNA-seq) technology allowed massively parallel characterization of thousands of cells at the transcriptome level and detected the heterogeneity of cell types in the tumor microenvironment [12,13]. Recently, scRNA-seq-induced drug repurposing improved accuracy and fidelity by using differentially expressed genes across different cell types [12]. For instance, researchers successfully utilized single-cell RNA sequencing to identify 19 different cell types in Bladder carcinoma (BC) environment and later highlighted the role of inflammatory cancer-associated fibroblasts (iCAFs) in tumor progression in BC, additionally providing essential data for future drug development [14]. For breast cancer, scRNA-seq was employed to separate and sequence breast cancer cells across three subtypes: Luminal (ER+, PR+/-), HER2+, and triple-negative (TNBC; ER-, PR-, HER2-). Researchers analyzed the data and, using scRNA-seq signatures, identified nine distinct "ecotypes" or clusters for breast cancer cells, each presenting unique cellular architectures [15]. Hence, by further applying the scRNA-seq technique, it was possible to predict the clinical outcomes of each ecotype cells based on its sequencing and structure, and to explore interactions with repurposed drugs. Thus, scRNA-seq permitted the detection of novel therapeutic targets, promoting drug repurposing studies for breast cancer [16].
This study conducts drug repurposing discovery for breast cancer based on scRNA-seq data. By analyzing the differentially expressed genes in different cell types of breast cancer, the study maps the genes to potential protein targets, followed by drug-target interaction analysis. This study provides a computational framework for the future design of combination treatment strategies for breast cancer therapies.
2. Methods and materials
2.1. Dataset
The scRNA-seq dataset for breast cancer was extracted from GEO database with accession ID: GSE158399. The original study was to obtain information about the lymph node metastasis of breast cancer cells, they selected the matched primary breast cancer (PC), positive lymph nodes (PL), and negative lymph nodes (NL) of the same patient to perform integrated analysis. The matched primary breast cancer (PC), positive lymph nodes (PL), and negative lymph nodes (NL) samples were analyzed with single-cell RNA sequencing.
2.2. KEGG
KEGG (Kyoto Encyclopedia of Genes and Genomes) is a knowledge database resource for understanding high-level functions and utilities of the biological system from molecular-level information, especially large-scale molecular datasets generated by genome sequencing and other high-throughput experimental technologies. The dataset is known to computerize data and knowledge on protein interaction networks (PATHWAY database) and to reconstruct protein interaction networks for all organisms whose genomes are completely sequenced (GENEsS and SSDB databases) [17]. This study merged KEGG dataset with the separated breast cancer single cell sets for further scRNA seq DEG analysis.
2.3. DrugBank
In this study, we used DrugBank datasets to obtain cancer-related drug candidates. DrugBank is a comprehensive, free-to-access, online database containing information on drugs and drug targets which combined detailed drug (chemical, pharmacological and pharmaceutical) data with comprehensive drug target (sequence, structure, and pathway) information [18]. repoDB, which is a database further developed for drug repurposing use, contains approved and failed drugs and their indications. It is the combination of datasets from both AACT and DrugCentral [19,20].
2.4. scRNA seq differentially expressed gene analysis
Single cell RNA sequenced differentially expressed gene analysis refers to the computation of the RNA molecules within each cell of a given sample and further analysis of that base on different gene expression [21]. Contrasting to the conventional bulk-RNA seq technique, scRNA-seq provides gene measurements for a genome wide range of individual cells and labels them based on the sorted information [22]. Thus, differentially expressed gene analysis under multiple conditions can be done with separated cell types, which allows direct comparison and biological interpretation for needs. One of the developed tools we used in this study is through pseudobulkDGE in scran [23]. It is a wrapper function around edgeR’s quasi-likelihood methods to conveniently perform differential expression analyses on pseudo-bulk profiles, allowing detection of cell type-specific changes between conditions in replicated studies.
2.5. Drug candidate generation
The generation and selection of drug candidates were crucial steps in this study. We needed to filter and identify cancer-related drugs that could effectively bind with protein targets for DTI docking. We utilized the Drug2cell tool, which identifies specific cellular targets of bioactive molecules with drug properties based on single-cell RNA-seq data. Drug2cell allows filtering of drugs and target molecules based on quantitative bioactivity metrics, drug categories (ATC classification), clinical trial phases, and classes of molecular targets. This approach reveals hidden mechanisms of action and predicts the impact of medicines on specific cell types [24].
2.6. Molecular featurization
We applied the molecular featurization on the drug candidates and proteins in order to the computational software to read and analyze the dataset. It plays a critical role in transforming the chemical structures of the drugs and gene sequences of proteins targets to feature vectors or numbers that can be passed down to learning algorithms since we required computational support to analyze the extensive dataset [25]. The developed tool we used in this project is RDKit, which is an open-source cheminformatics and machine learning machine [26]. The RDKit supports several different models and allows us to define our own by providing a function that assigns aromaticity.
2.7. Molecular docking simulation
We used the molecular docking methodology to explore the behavior of small molecules (drugs) in the binding site of a target protein. Because it is crucial in predicting the orientation of the ligand/drug when it is bound to a protein receptor using shape and electrostatic interactions to quantify it in modern drug discovery [27]. The docking process involves two steps: prediction of the ligand conformation, position, and orientation within these sites (usually referred to as pose) and assessment of the binding affinity. Autodock vina, a docking engine we used in the project, only requires the structures of the molecules being docked and the specification of the search space including the binding site [28].
3. Results
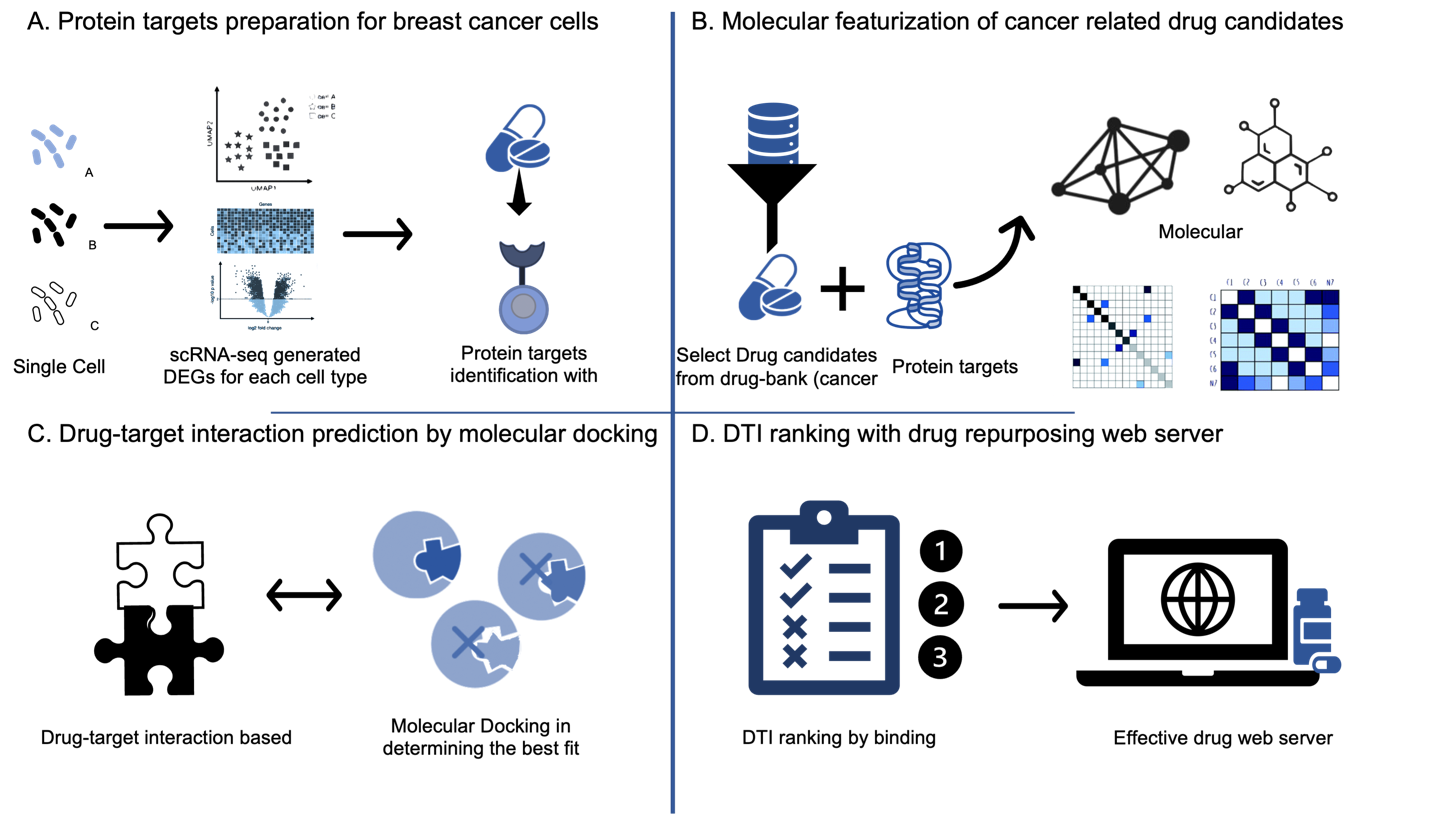
Figure 1. Framework of drug repurposing by single cell RNA-seq for breast cancer. Panel A shows the protein targets preparation for breast cancer cells through scRNA-seq analysis following by the mapping of DEGs into correlated protein targets through Uniprot database. Panel B shows the molecular featurization of cancer related drug candidates from drug bank based on its chemical structures and prior clinical trial results. Panel C shows drug-target interaction prediction by molecular docking (Autodock Vina), which first predict the protein-ligand conformation, and secondly assess the binding affinity. Panel D shows the DTI ranking followed by the integration of all the computational pipelines and dataset to establish an efficient drug repurposing protocol for breast cancer.
3.1. Framework of drug repurposing by single cell RNA-seq for breast cancer
Figure 1 shows the framework of drug repurposing for breast cancer. First, we collected and clustered various types of breast cancer cells based on their genomic characteristics. We then performed scRNA-seq statistical analysis to generate a list of differentially expressed genes (DEGs) for each cell type. These DEGs were analyzed through univariate and multivariate methods and mapped to corresponding protein targets using the UniProt database. Next, we filtered and selected candidate cancer-related drugs after identifying the DEGs and their associated protein targets. Using molecular docking simulations with AutoDock Vina, we examined the behavior of small molecules (drugs) at the binding sites of target proteins. The best-fit candidate drugs for each cell-type protein target were ranked based on binding scores, resulting in a list of the top repurposed drugs. Finally, we integrated all computational pipelines and datasets to establish an efficient drug repurposing web server for breast cancer.
3.2. Single-cell RNA sequencing data analysis
After processing the breast cancer scRNA-seq data, we annotated ten cell types: Pericytes, Myeloid cells, CD8 Effector cells, B cells, Myofibroblasts, Naive T cells, CXCL14 cancer cells, Plasma cells, Macrophages, and Cancer stem cells (Figure 2). Among these, Pericytes and Myeloid cells had the most drug candidates applied to them. Differentially expressed genes (DEGs) were identified through pseudobulk DEG analysis in Scran, considering cell fractions and mean expression levels within groups. Key DEGs included CACNA1I, MS4A1, COL5A2, COL6A3, COL3A1, IL12B, PDE7B, COL4A1, and IL5RA. By using these top ten DEGs, we identified over a hundred protein targets. For example, Q01344 and P11836 are cell membrane proteins that exhibit specific linkage with HER2+ breast cancer subtype,[29]. and Q9P0X4 is associated with the TNBC subtype.
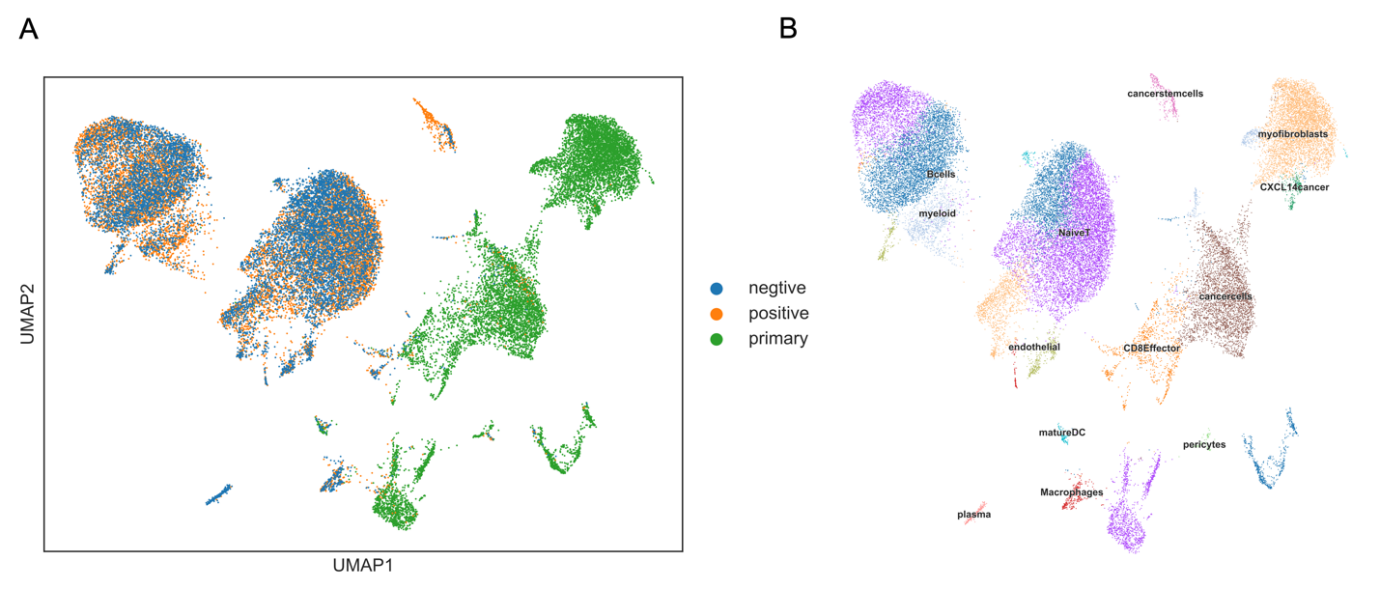
Figure 2. Leiden clustering and cell type annotation of scRNA-seq data in three subtypes of breast cancer. Panel A shows the Leiden clustering of scRNA-seq data. Panel B shows the cell type annotation of scRNA-seq data in breast cancer.
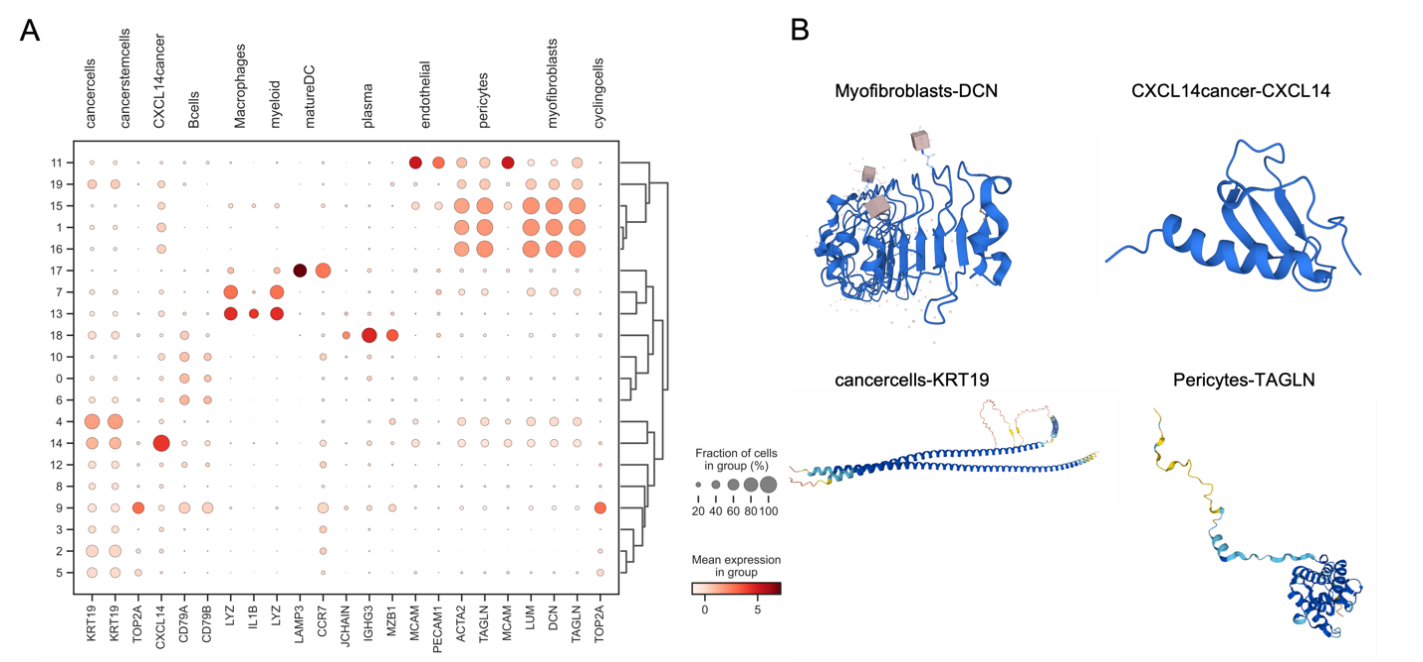
Figure 3. Differentially expressed gene analysis in scRNA-seq data and examples of drug candidates. Panel A shows the differentially expressed gene analysis in scRNA-seq data. Panel B shows the examples of drug candidates across different cell types.
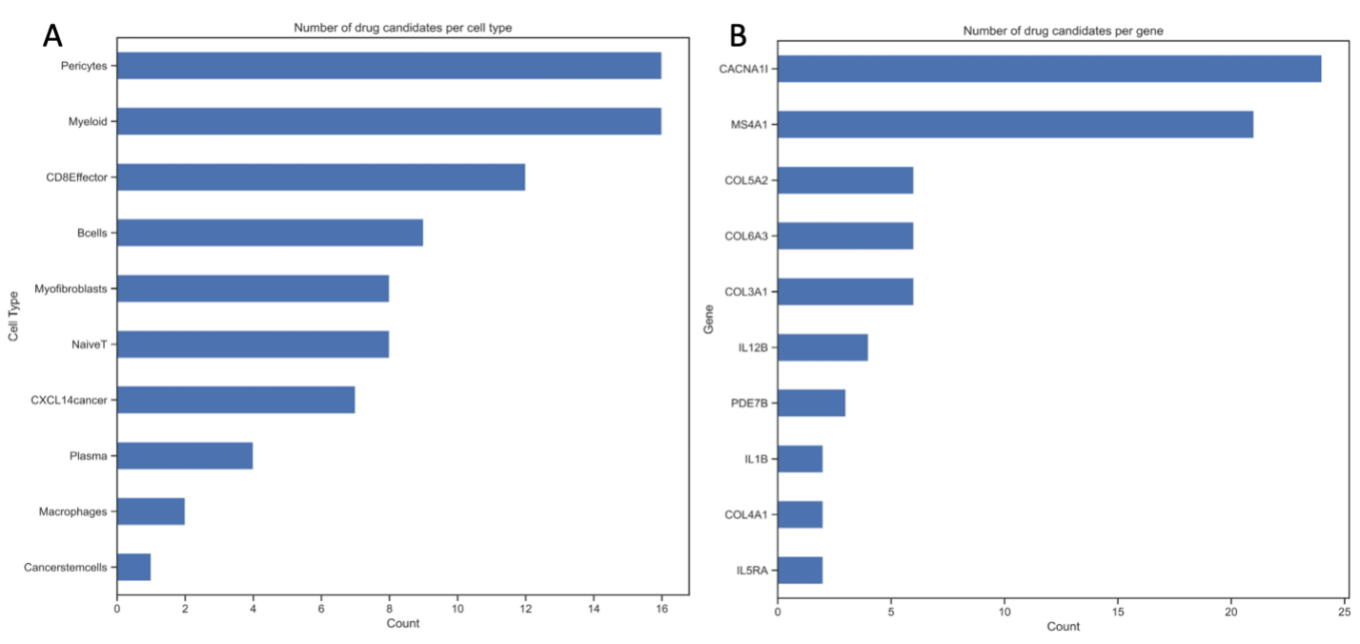
Figure 4. The number of drug candidates applied on each target. Panel A shows the numbers of drug candidates found per cell type. Panel B shows the numbers of drug candidates found per gene. This showcases the cell types and genes that are most suitable for breast cancer and repurposed drugs targeting.
3.3. Drug repurposing by drug2cell and molecular docking simulations
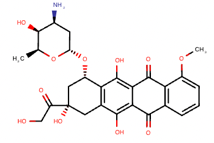
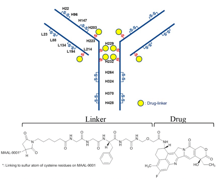
Based on the DEGs, drug candidates were selected using the Drug2cell (Figure 3). These candidates include Doxorubicin, Cyclophosphamide, Everolimus, Tamoxifen, Anastrozole, Paclitaxel, Aspirin, Trastuzumab deruxtecan, Capivasertib, Gemcitabine, Methotrexate, Itraconazole, Simvastatin, and Metformin (Table 1). For all candidates, we also show the number of drug candidates applied on each target in Figure 4. The proteins and drugs were processed through RDKit and AutoDock Vina for sorting and scoring their drug-target interactions (Figure 5). Among the candidates, fourteen drugs were particularly notable, including Doxorubicin, Cyclophosphamide, and Everolimus for the HER2+ cancer subtype, and Paclitaxel and Gemcitabine for TNBC subtypes. These drugs were repurposed with clinical evidence supporting their effectiveness in breast cancer treatment.
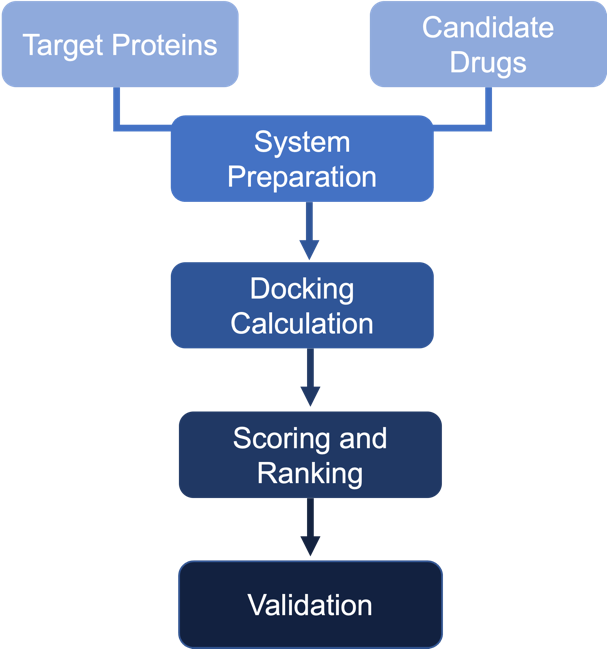
Figure 5. Flow chart showing the processes of drug-target interaction (DTI).
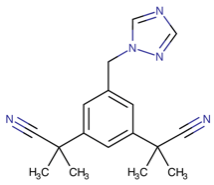
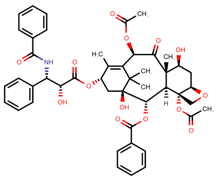
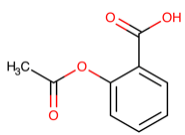
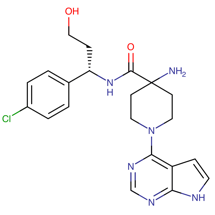
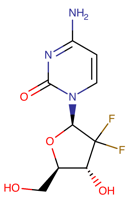
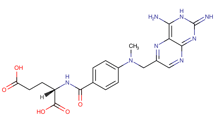
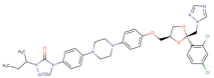
Table 1. The top candidates for feasible repurposed drugs with the related chemical structures, biomarkers, responsible breast cancer types for treatments, and reported clinical outcomes.
Drug | Structure | Biomarkers | Breast cancer type | Reported Outcomes |
Doxorubicin | DB00997
| CREB3L1 HLA region | “Docetaxel, and ERBB2+ and basal-like subtypes” = TNBC | Tumor sensitivity; Cardiotoxicity predisposition |
Cyclophosphamide | DB00531
| CYP2B6 and CYP2C19 | HER2-Positive | Drug metabolic rate |
Everolimus | DB01590
| CYP3A4 ABCB1 PI3KR1 RAPTOR | Advanced hormone receptor-positive, HER2-negative (advanced HR+ BC) | Higher plasma concentration of Everolimus Adverse Side Effects |
Tamoxifen | DB00675
| CYP2D6 CYPI9A1 | Estrogen (hormone) receptor-positive Luminal A | Metabolic rate. Effectiveness of the drugs |
Anastrozole | DB01217
| CSMD1 CYP19A1 | Hormone receptor-positive (ER) | Increased anastrozole sensitivity Drug response |
Paclitaxel | DB01229
| SNPs on LPHN2, R0B01, SNTG1 and GRIk1 | ER-positive TNBC | Insensitivity to drug. Bad prognosis |
Aspirin | DB00945
| PIK3CA mutation | Hormone Receptor-positive | Drug Efficacy1 |
Trastuzumab deruxtecan | DB14962
| HER2-positive2 | HER2-positive | Gene expression |
Capivasertib | DB12218
| PIK3CA mutation PTEN loss3 AKT Activation4 | Hormone receptor-positive HER2-positive | Drug Efficacy Drug response |
Gemcitabine | DB00441
| BRCA mutation5 ER, PR, and HER2 | TNBC (basal like) | Increased sensitivity to drug |
Methotrexate | DB00563
| ER, PR, and HER26 | TNBC | Drug response |
Itraconazole | DB01167
| SKBR3 &MCF7 | TNBC | Decreased tumor size |
Simvastatin | DB00641
| ER, PR, HER27 Ki-67 BRCA mutation PTTG1 Gene | TNBC | Less cell proliferation Drug Efficacy |
Metformin | DB00331
| pAMPK8 Ki-67 | ER-positive | Down regulation Drug response cascade |
4. Conclusions and future directions
Although the fourteen drugs exhibited high efficiency in treating breast cancer, some clinical trials revealed some adverse effects on other body tissues, such as over-toxicity and apoptosis of healthy cells [30]. Despite these challenges, the drug-repurposing strategy combined with scRNA-seq data has the potential to enhance therapeutic target discovery and improve treatment success rates for breast cancer.
References
[1]. Antfolk, M., Kim, S., Koizumi, S. et al. Label-free single-cell separation and imaging of cancer cells using an integrated microfluidic system. Sci Rep 7, 46507 (2017).
[2]. Sun YS, Zhao Z, Yang ZN, Xu F, Lu HJ, Zhu ZY, Shi W, Jiang J, Yao PP, Zhu HP. Risk Factors and Preventions of Breast Cancer. Int J Biol Sci. 2017 Nov 1;13(11):1387-1397. doi: 10.7150/ijbs.21635. PMID: 29209143; PMCID: PMC5715522.
[3]. Sharma GN, Dave R, Sanadya J, Sharma P, Sharma KK. Various types and management of breast cancer: an overview. J Adv Pharm Technol Res. 2010 Apr;1(2):109-26. PMID: 22247839; PMCID: PMC3255438.
[4]. Global Burden of Disease Cancer Collaboration. et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2016: a systematic analysis for the Global Burden of Disease study. JAMA Oncol. 4, 1553–1568 (2018).
[5]. Arnold M, Morgan E, Rumgay H, Mafra A, Singh D, Laversanne M, Vignat J, Gralow JR, Cardoso F, Siesling S, Soerjomataram I. Current and future burden of breast cancer: Global statistics for 2020 and 2040. Breast. 2022 Dec;66:15-23. doi: 10.1016/j.breast.2022.08.010. Epub 2022 Sep 2. PMID: 36084384; PMCID: PMC9465273.
[6]. Skipper, H.E. (1978), Adjuvant chemotherapy. Cancer, 41: 936-940.
[7]. Ahmad Malik, Jonaid, et al. ‘Breast Cancer Drug Repurposing a Tool for a Challenging Disease’. Drug Repurposing - Molecular Aspects and Therapeutic Applications, IntechOpen, 1 June 2022. Crossref, doi:10.5772/intechopen.101378.
[8]. Chung SS, Dutta P, Chard N, Wu Y, Chen QH, Chen G, Vadgama J. A novel curcumin analog inhibits canonical and non-canonical functions of telomerase through STAT3 and NF-κB inactivation in colorectal cancer cells. Oncotarget. 2019 Jul 16;10(44):4516-4531. doi: 10.18632/oncotarget.27000. PMID: 31360301; PMCID: PMC6642039.
[9]. Liu Z, Li Q, Li K, Chen L, Li W, Hou M, Liu T, Yang J, Lindvall C, Björkholm M, Jia J, Xu D. Telomerase reverse transcriptase promotes epithelial-mesenchymal transition and stem cell-like traits in cancer cells. Oncogene. 2013
[10]. Pan, X, Lin, X, Cao, D, Zeng, X, Yu, PS, He, L, et al. Deep learning for drug repurposing: Methods, databases, and applications. WIREs Comput Mol Sci. 2022; 12:e1597.
[11]. Schmidt DR, Patel R, Kirsch DG, Lewis CA, Vander Heiden MG, Locasale JW. Metabolomics in cancer research and emerging applications in clinical oncology. CA Cancer J Clin. 2021 Jul;71(4):333-358. doi: 10.3322/caac.21670. Epub 2021 May 13. PMID: 33982817; PMCID: PMC8298088.
[12]. Mohammadi, E., Jin, H., Zhang, C., Shafizade, N., Dashty, S., Lam, S., ... & Sekhavati, M. H. (2022). Drug repositioning for immunotherapy in breast cancer using single-cell and spatial transcriptomics analysis. medRxiv, 2022-11.
[13]. Kumar, Manu P., et al. "Analysis of single-cell RNA-seq identifies cell-cell communication associated with tumor characteristics." Cell reports 25.6 (2018): 1458-1468.
[14]. Chen, Z., Zhou, L., Liu, L. et al. Single-cell RNA sequencing highlights the role of inflammatory cancer-associated fibroblasts in bladder urothelial carcinoma. Nat Commun 11, 5077 (2020). https://doi.org/10.1038/s41467-020-18916-5
[15]. Wu, S. Z., Al-Eryani, G., Roden, D. L., Junankar, S., Harvey, K., Andersson, A., ... & Swarbrick, A. (2021). A single-cell and spatially resolved atlas of human breast cancers. Nature genetics, 53(9), 1334-1347.
[16]. Ren L, Li J, Wang C, Lou Z, Gao S, Zhao L, Wang S, Chaulagain A, Zhang M, Li X, Tang J. Single cell RNA sequencing for breast cancer: present and future. Cell Death Discov. 2021 May 14;7(1):104. doi: 10.1038/s41420-021-00485-1. PMID: 33990550; PMCID: PMC8121804.
[17]. Ogata, H., Goto, S., Sato, K., Fujibuchi, W., Bono, H., & Kanehisa, M. (1999). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic acids research, 27(1), 29-34.
[18]. Wishart, D. S., Knox, C., Guo, A. C., Cheng, D., Shrivastava, S., Tzur, D., ... & Hassanali, M. (2008). DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic acids research, 36(suppl_1), D901-D906.
[19]. Brown, Adam S., and Chirag J. Patel. "A standard database for drug repositioning." Scientific data 4.1 (2017): 1-7.
[20]. Xue, H., Li, J., Xie, H., & Wang, Y. (2018). Review of drug repositioning approaches and resources. International journal of biological sciences, 14(10), 1232
[21]. Chen, G., Ning, B., & Shi, T. (2019). Single-cell RNA-seq technologies and related computational data analysis. Frontiers in genetics, 10, 317.
[22]. Li, X., & Wang, C. Y. (2021). From bulk, single-cell to spatial RNA sequencing. International Journal of Oral Science, 13(1), 36.
[23]. Younes, S. T., Showmaker, K., Johnson, A. C., Garrett, M. R., & Ryan, M. J. (2021). Single cell RNA sequencing reveals ferritin as a key mediator of autoimmune pre-disposition in a mouse model of systemic lupus erythematosus. Scientific Reports, 11(1), 24245.
[24]. Kanemaru, Kazumasa, Cranley, James, Muraro, Daniele, Miranda, Antonio M. A., Ho, Siew Yen et al. Nature 2023;619;7971;801
[25]. Wu, Z., Ramsundar, B., Feinberg, E. N., Gomes, J., Geniesse, C., Pappu, A. S., ... & Pande, V. (2018). MoleculeNet: a benchmark for molecular machine learning. Chemical science, 9(2), 513-530.
[26]. Bento, A. P., Hersey, A., Félix, E., Landrum, G., Gaulton, A., Atkinson, F., ... & Leach, A. R. (2020). An open source chemical structure curation pipeline using RDKit. Journal of Cheminformatics, 12, 1-16.
[27]. Meng XY, Zhang HX, Mezei M, Cui M. Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des. 2011 Jun;7(2):146-57. doi: 10.2174/157340911795677602. PMID: 21534921; PMCID: PMC3151162.
[28]. Pagadala NS, Syed K, Tuszynski J. Software for molecular docking: a review. Biophys Rev. 2017 Apr;9(2):91-102. doi: 10.1007/s12551-016-0247-1. Epub 2017 Jan 16. PMID: 28510083; PMCID: PMC5425816.
[29]. Yang V, Gouveia MJ, Santos J, Koksch B, Amorim I, Gärtner F, Vale N. Breast cancer: insights in disease and influence of drug methotrexate. RSC Med Chem. 2020 May 28;11(6):646-664. doi: 10.1039/d0md00051e. PMID: 33479665; PMCID: PMC7578709.
[30]. Royce ME, Osman D. Everolimus in the Treatment of Metastatic Breast Cancer. Breast Cancer (Auckl). 2015 Sep 6;9:73-9. doi: 10.4137/BCBCR.S29268. PMID: 26417203; PMCID: PMC4571987.
Cite this article
Zhao,J. (2024). Cell type specific drug repurposing for breast cancer by integrating single-cell RNA sequencing and molecular docking simulation. Theoretical and Natural Science,49,233-242.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 4th International Conference on Biological Engineering and Medical Science
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Antfolk, M., Kim, S., Koizumi, S. et al. Label-free single-cell separation and imaging of cancer cells using an integrated microfluidic system. Sci Rep 7, 46507 (2017).
[2]. Sun YS, Zhao Z, Yang ZN, Xu F, Lu HJ, Zhu ZY, Shi W, Jiang J, Yao PP, Zhu HP. Risk Factors and Preventions of Breast Cancer. Int J Biol Sci. 2017 Nov 1;13(11):1387-1397. doi: 10.7150/ijbs.21635. PMID: 29209143; PMCID: PMC5715522.
[3]. Sharma GN, Dave R, Sanadya J, Sharma P, Sharma KK. Various types and management of breast cancer: an overview. J Adv Pharm Technol Res. 2010 Apr;1(2):109-26. PMID: 22247839; PMCID: PMC3255438.
[4]. Global Burden of Disease Cancer Collaboration. et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 cancer groups, 1990 to 2016: a systematic analysis for the Global Burden of Disease study. JAMA Oncol. 4, 1553–1568 (2018).
[5]. Arnold M, Morgan E, Rumgay H, Mafra A, Singh D, Laversanne M, Vignat J, Gralow JR, Cardoso F, Siesling S, Soerjomataram I. Current and future burden of breast cancer: Global statistics for 2020 and 2040. Breast. 2022 Dec;66:15-23. doi: 10.1016/j.breast.2022.08.010. Epub 2022 Sep 2. PMID: 36084384; PMCID: PMC9465273.
[6]. Skipper, H.E. (1978), Adjuvant chemotherapy. Cancer, 41: 936-940.
[7]. Ahmad Malik, Jonaid, et al. ‘Breast Cancer Drug Repurposing a Tool for a Challenging Disease’. Drug Repurposing - Molecular Aspects and Therapeutic Applications, IntechOpen, 1 June 2022. Crossref, doi:10.5772/intechopen.101378.
[8]. Chung SS, Dutta P, Chard N, Wu Y, Chen QH, Chen G, Vadgama J. A novel curcumin analog inhibits canonical and non-canonical functions of telomerase through STAT3 and NF-κB inactivation in colorectal cancer cells. Oncotarget. 2019 Jul 16;10(44):4516-4531. doi: 10.18632/oncotarget.27000. PMID: 31360301; PMCID: PMC6642039.
[9]. Liu Z, Li Q, Li K, Chen L, Li W, Hou M, Liu T, Yang J, Lindvall C, Björkholm M, Jia J, Xu D. Telomerase reverse transcriptase promotes epithelial-mesenchymal transition and stem cell-like traits in cancer cells. Oncogene. 2013
[10]. Pan, X, Lin, X, Cao, D, Zeng, X, Yu, PS, He, L, et al. Deep learning for drug repurposing: Methods, databases, and applications. WIREs Comput Mol Sci. 2022; 12:e1597.
[11]. Schmidt DR, Patel R, Kirsch DG, Lewis CA, Vander Heiden MG, Locasale JW. Metabolomics in cancer research and emerging applications in clinical oncology. CA Cancer J Clin. 2021 Jul;71(4):333-358. doi: 10.3322/caac.21670. Epub 2021 May 13. PMID: 33982817; PMCID: PMC8298088.
[12]. Mohammadi, E., Jin, H., Zhang, C., Shafizade, N., Dashty, S., Lam, S., ... & Sekhavati, M. H. (2022). Drug repositioning for immunotherapy in breast cancer using single-cell and spatial transcriptomics analysis. medRxiv, 2022-11.
[13]. Kumar, Manu P., et al. "Analysis of single-cell RNA-seq identifies cell-cell communication associated with tumor characteristics." Cell reports 25.6 (2018): 1458-1468.
[14]. Chen, Z., Zhou, L., Liu, L. et al. Single-cell RNA sequencing highlights the role of inflammatory cancer-associated fibroblasts in bladder urothelial carcinoma. Nat Commun 11, 5077 (2020). https://doi.org/10.1038/s41467-020-18916-5
[15]. Wu, S. Z., Al-Eryani, G., Roden, D. L., Junankar, S., Harvey, K., Andersson, A., ... & Swarbrick, A. (2021). A single-cell and spatially resolved atlas of human breast cancers. Nature genetics, 53(9), 1334-1347.
[16]. Ren L, Li J, Wang C, Lou Z, Gao S, Zhao L, Wang S, Chaulagain A, Zhang M, Li X, Tang J. Single cell RNA sequencing for breast cancer: present and future. Cell Death Discov. 2021 May 14;7(1):104. doi: 10.1038/s41420-021-00485-1. PMID: 33990550; PMCID: PMC8121804.
[17]. Ogata, H., Goto, S., Sato, K., Fujibuchi, W., Bono, H., & Kanehisa, M. (1999). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic acids research, 27(1), 29-34.
[18]. Wishart, D. S., Knox, C., Guo, A. C., Cheng, D., Shrivastava, S., Tzur, D., ... & Hassanali, M. (2008). DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic acids research, 36(suppl_1), D901-D906.
[19]. Brown, Adam S., and Chirag J. Patel. "A standard database for drug repositioning." Scientific data 4.1 (2017): 1-7.
[20]. Xue, H., Li, J., Xie, H., & Wang, Y. (2018). Review of drug repositioning approaches and resources. International journal of biological sciences, 14(10), 1232
[21]. Chen, G., Ning, B., & Shi, T. (2019). Single-cell RNA-seq technologies and related computational data analysis. Frontiers in genetics, 10, 317.
[22]. Li, X., & Wang, C. Y. (2021). From bulk, single-cell to spatial RNA sequencing. International Journal of Oral Science, 13(1), 36.
[23]. Younes, S. T., Showmaker, K., Johnson, A. C., Garrett, M. R., & Ryan, M. J. (2021). Single cell RNA sequencing reveals ferritin as a key mediator of autoimmune pre-disposition in a mouse model of systemic lupus erythematosus. Scientific Reports, 11(1), 24245.
[24]. Kanemaru, Kazumasa, Cranley, James, Muraro, Daniele, Miranda, Antonio M. A., Ho, Siew Yen et al. Nature 2023;619;7971;801
[25]. Wu, Z., Ramsundar, B., Feinberg, E. N., Gomes, J., Geniesse, C., Pappu, A. S., ... & Pande, V. (2018). MoleculeNet: a benchmark for molecular machine learning. Chemical science, 9(2), 513-530.
[26]. Bento, A. P., Hersey, A., Félix, E., Landrum, G., Gaulton, A., Atkinson, F., ... & Leach, A. R. (2020). An open source chemical structure curation pipeline using RDKit. Journal of Cheminformatics, 12, 1-16.
[27]. Meng XY, Zhang HX, Mezei M, Cui M. Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des. 2011 Jun;7(2):146-57. doi: 10.2174/157340911795677602. PMID: 21534921; PMCID: PMC3151162.
[28]. Pagadala NS, Syed K, Tuszynski J. Software for molecular docking: a review. Biophys Rev. 2017 Apr;9(2):91-102. doi: 10.1007/s12551-016-0247-1. Epub 2017 Jan 16. PMID: 28510083; PMCID: PMC5425816.
[29]. Yang V, Gouveia MJ, Santos J, Koksch B, Amorim I, Gärtner F, Vale N. Breast cancer: insights in disease and influence of drug methotrexate. RSC Med Chem. 2020 May 28;11(6):646-664. doi: 10.1039/d0md00051e. PMID: 33479665; PMCID: PMC7578709.
[30]. Royce ME, Osman D. Everolimus in the Treatment of Metastatic Breast Cancer. Breast Cancer (Auckl). 2015 Sep 6;9:73-9. doi: 10.4137/BCBCR.S29268. PMID: 26417203; PMCID: PMC4571987.