1. Introduction
The advent of CRISPR-Cas systems has revolutionized genetic engineering, offering unprecedented precision and ease in manipulating DNA sequences. CRISPR (clustered regularly interspaced short palindromic repeats) and Cas (CRISPR-associated) proteins were initially discovered as part of bacteria's adaptive immune system, where they protect against invading genetic elements such as viruses and plasmids [1]. The system's potential for genome editing was first recognized in 2012 when researchers demonstrated that Cas9, a protein from Streptococcus pyogenes, could be repurposed to target and cleave specific DNA sequences in a programmable manner [1]. This discovery opened the floodgates to myriad applications, from basic research in molecular biology to therapeutic interventions in human diseases.
CRISPR-Cas systems have evolved into a suite of tools enabling researchers to perform a variety of genetic modifications, including gene knockouts, insertions, and precise edits [2]. The simplicity and versatility of the CRISPR-Cas9 system have been pivotal in its widespread adoption. Unlike earlier gene-editing technologies such as zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), CRISPR-Cas9 does not require complex protein engineering for each new target sequence. Instead, a small guide RNA (gRNA) directs the Cas9 nuclease to the desired genomic location, enabling rapid and cost-effective gene targeting [2]. This modularity has made CRISPR-Cas9 a powerful tool for scientists, facilitating studies that were previously infeasible due to technical or economic constraints.
Despite its remarkable capabilities, the CRISPR-Cas system is not without challenges. Concerns about off-target effects, where unintended genomic sites are modified, have been a major focus of ongoing research [3]. The potential for such off-target effects can lead to unpredictable consequences, including the disruption of essential genes or the activation of oncogenes. Addressing these issues is crucial for the safe and effective application of CRISPR-Cas technologies, particularly in clinical settings. Recent advancements have focused on improving the specificity and fidelity of the CRISPR-Cas system through the development of high-fidelity Cas9 variants and alternative Cas proteins, such as Cas12a and Cas13 [4]. These innovations aim to minimize off-target effects while maintaining the high efficiency that has made CRISPR-Cas a cornerstone of modern molecular biology.
2. Different Types of CRISPR-Cas Systems
An adaptive immune system, known as the CRISPR-Cas system, is widespread among bacteria and archaea [5]. This revolutionary gene-editing technology utilizes bacterial defense mechanisms to precisely locate and cut DNA sequences, facilitating gene modification or deletion. Its emergence has significantly advanced life science research, leading to breakthroughs in disease treatment and agricultural biotechnology. In response to this bacterial immune system, phages have evolved various mechanisms to counteract CRISPR-Cas, with one efficient method being the production of anti-CRISPR proteins (Acrs) to disable the system. The CRISPR-Cas system is broadly classified into two categories [6]: Class I (including types I, III, and IV), which utilize multiple Cas proteins and constitute 90% of identified CRISPR-Cas systems; and Class II (including Type II/Cas9, Type V/Cas12, and Type VI/Cas13), which function via a single Cas protein and are particularly useful for gene editing and in vitro assays. Among these, Type II, employing the Cas9 protein, is the most widely used.
Studies have shown that the CRISPR-Cas9 system is the most widely used genome editing technology, with broad applications in gene function research, model animal construction, and gene therapy for conditions such as cancer [6], liver disease, and cardiovascular disease [7]. The CRISPR-Cas9 system consists primarily of Cas9 and sgRNA, formed by tracrRNA and crRNA. Its mechanism relies on RNA-guided DNA cleavage and repair, where the crRNA and tracrRNA combine to recognize and bind to the target sequence, forming a complex. Subsequently, the Cas9 protein, guided by the crRNA, initiates DNA cleavage, triggering the target DNA's repair mechanism and enabling gene editing [8]. CRISPR-Cas9-mediated gene integration predominantly occurs via homologous recombination and non-homologous recombination. However, the use of Cas9 for DNA cleavage necessitates a vector and may lead to off-target effects, underscoring the importance of carefully selecting target sequences to minimize experimental risks [9].
3. Toolalization of native CRISPR with the design of sgRNA
The sgRNA is an essential component of gene editing technology, involving the design, optimization, and evaluation of sgRNA. Utilizing sgRNA instrumentation ensures precise and efficient guidance of the Cas9 protein for gene editing, thereby enhancing the systematic and automated nature of the process. This improvement, in turn, enhances the accuracy and efficiency of gene editing [10]. Additionally, sgRNA tools contribute significantly to the dissemination and application of gene editing technology, providing robust support for developing genome-wide programmable transcriptional memory in biomedical research and other fields (such as CRISPR-based epigenome editing) [11]. The use of sgRNA design tools facilitates the identification of optimal sgRNAs for experiments. However, a major challenge in effectively applying the CRISPR system is accurately predicting the efficacy of sgRNAs for targeted knockdown and their potential off-target effects.
The precision of sgRNA predictions is crucial for designing sgRNAs that are both efficient and highly specific, directly influencing the accuracy and reliability of CRISPR technology in gene editing. Therefore, optimizing the sgRNA design process to guide Cas proteins accurately and efficiently in editing target DNA sequences while minimizing off-target effects at non-target sites is essential for advancing CRISPR technology [12]. The optimization of sgRNAs holds a significant position in gene editing, with previous researchers contributing substantially to developing computational design rules and evaluation criteria for optimized sgRNAs [10]. The evaluation of sgRNAs plays a pivotal role in the final screening of gene editing guide RNAs, acting as a gatekeeper in the selection process. Various software packages, such as ZiFiT [13], E-CRISP [14], and CCTop [15], have been developed for designing and evaluating sgRNAs [16], underscoring the importance of meticulous attention to each step in our research on sgRNA instrumentation for gene editing.
The field of sgRNA design has made significant advancements, playing a crucial role in CRISPR-Cas9 gene editing technology. Current priorities in sgRNA design include enhancing specificity, reducing off-target effects, and improving editing efficiency [17]. Several popular sgRNA design tools are available, such as CRISPOR [18] and the GPP Web Portal, with other notable options including E-CRISP [14] and CHOPCHOP [19]. Specificity in sgRNA design is critical, as similarity to other gene sequences can lead to off-target editing. Off-target effects occur when sgRNA mismatches are recognized, potentially causing chromosomal rearrangements and genomic damage, limiting their therapeutic application. Moreover, off-target effects may disrupt gene function, leading to various physiological or signaling abnormalities [20]. To mitigate these risks, it is advisable to select target sequences that are distinct from other gene sequences.
In the process of designing sgRNAs, the first step is to determine the target sequence, which corresponds to the sequence of the gene intended for editing. The selection of the target sequence should prioritize factors such as complementarity, specificity, and the presence of a PAM sequence (Protospacer Adjacent Motif). The PAM sequence is crucial as it enables the Cas9 protein to recognize and bind to the target sequenc . Next, sequence analysis tools are employed to identify the target sequence. Following this, the sgRNA is designed, comprising two components: crRNA (CRISPR RNA) and tracrRNA (trans-activating crRNA). The crRNA is responsible for complementary base pairing with the target DNA sequence, while tracrRNA interacts with the Cas9 protein to form a functional complex. These two RNA components can be synthesized into a single molecule known as sgRNA. Additionally, the base composition of sgRNA must be considered. It is recommended to avoid sequences ending with more than four consecutive T bases, and the optimal GC% content ranges from 30% to 70% (preferably 40% to 60%). Ideally, sgRNA should start with a G base at its 5' end, or an artificial G can be added to enhance transcription efficiency. Evaluation of potential off-target effects and consideration of editing efficiency are also crucial steps. Finally, an expression vector capable of expressing sgRNA must be constructed. In conclusion, the design of sgRNA involves comprehensive consideration of multiple factors, ensuring the construction of well-designed sgRNA facilitates efficient and precise gene editing.
4. Applications of CRISPR-Cas System
The genome editing toolbox is rapidly expanding thanks to the discovery of novel, easy-to-use, and versatile tools, most notably the CRISPR-Cas system, an acquired immunity system for prokaryotes to resist the invasion of exogenous genetic elements such as phages or plasmids. The system consists of two core components, the CRISPR sequence and the Cas gene, which can accurately recognize and cut off exogenous nucleic acids and silence the expression of exogenous genes, thus maintaining the stability of its genetic system [21-23]. The CRISPR-Cas system has a wide range of applications, and its potential is gradually being explored and applied in many cutting-edge fields, it has achieved partial success in experimental tools, cells, eukaryotes, and gene therapy. part of its success in experimental tools, cells, eukaryotes, and gene therapy.
The CRISPR-Cas9 system(Figure 1 Mechanism of action of the CRISPR-Cas 9 system)has proven effective in disrupting and modifying genes, demonstrating significant potential for cancer treatment. It has become one of the most promising tools in basic biomedical research and therapeutic applications. CRISPR-Cas9-based gene editing technology can potentially cure diseases by targeting mutated genes, knocking out proto-oncogenes, and developing drugs to treat malignant tumors caused by genetic variations [24]. Engineering human T cells for treating cancer, viral infections, and autoimmunity has long been a goal for immunologists and hematologists. The adaptation of CRISPR-Cas9 to human T cells has made T cell engineering techniques widely accessible and has accelerated the development of engineered T cell therapies. Novel Cas proteins, both natural and engineered, are rapidly emerging, offering increased flexibility, activity, and specificity. Advanced protein engineering techniques, such as fusing Cas proteins with deaminases or reverse transcriptases, enable genomic DNA editing without the need for double-stranded cleavage. As a result, the "CRISPR toolbox" for experimental use and novel therapeutic approaches is rapidly expanding [25]. Genetic manipulation holds profound implications for both preclinical and clinical research.
The CRISPR-Cas system has revolutionized the field of genome editing by providing a simple and inexpensive way to target genomes. In recent years, the ease of targeting eukaryotic DNA with the CRISPR-Cas system has "democratized" therapeutic genome editing research and facilitated the development of promising gene editing technologies for many genetic diseases [26]. These include the first-generation gene editing technology, zinc-finger nuclease technology, the second-generation gene editing technology, transcription activator-like effector nuclease technology, and the third-generation gene editing technology, the CRISPR-Cas system. Compared to the first two generations of technology, the CRISPR-Cas system is characterized by higher targeting accuracy, easier design and construction, higher cleavage efficiency, and less cytotoxicity. Additionally, the CRISPR-Cas system can realize multi-gene editing, demonstrating great potential for application. Currently, CRISPR-Cas technology has become a research hotspot in the field of gene editing and is expected to dominate the future development of gene editing technology [27]. Despite the rapid advancements in gene editing technology in recent years, it is important to note that using gene editing technology on humans is considered unethical.
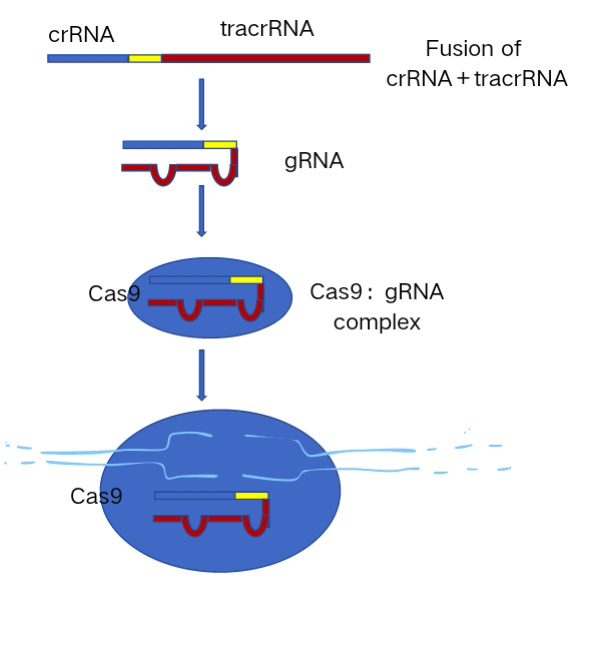
Figure 1. Mechanism of action of the CRISPR-Cas9 system after instrumentation.
5. Discussion/Perspectives
The CRISPR-Cas system, a revolutionary gene-editing tool, cleverly draws on the bacterial immune mechanism of nature and enables fine manipulation of DNA. The second class of members of the family, such as Cas 9 and Cas 12, attracts much attention for their independent protein functions and mild properties. At the heart of the technology forward is optimizing the design strategy for sgRNA (single-stranded oriented RNA). CRISPR-Cas technology has been widely penetrated into the forefront of experimental science, cell exploration, eukaryotic research and gene therapy, especially in the infinite potential of anti-cancer journey. Compared with traditional technologies, CRISPR / Cas system stands out with its excellent targeting accuracy, construction convenience, high efficiency and low toxicity, and has the ability to edit multiple genes at the same time, laying a solid foundation for the future blueprint of gene editing. As an emerging technology, I believe that the CRISPR-Cas system has great potential, as a cutting-edge gene editing technology, its prospect is very broad. In the scientific research and exploration, CRISPR-Cas has revolutionized the way of gene function research and disease mechanism analysis by virtue of its extraordinary efficiency, accuracy and programmability. With the increasing improvement of technology, it is gradually becoming an important engine to promote the leap of basic science. In the field of health care, this system provides unprecedented innovative therapies for the treatment of genetic diseases and cancer, by accurately repairing or replacing defective genes, and steadily moving towards the goal of cure. At the same time, CRISPR-Cas has shown broad application prospects in vaccine development, drug screening and even broader medical practice. In addition, its influence also extends to the field of agricultural biotechnology and biosynthesis. Through accurate gene editing, it helps to cultivate high-yield and stress-resistant crop varieties, leading the green reform of agricultural production. In addition, the technology can also synthesize customized biomaterials, bringing breakthrough solutions to cutting-edge fields such as biomedical research and bio-energy development, demonstrating its great value across industries and fields. In conclusion, the CRISPR-Cas system has wide application prospects and great development potential, and will play an increasingly important role in the fields of life sciences and medicine.
References
[1]. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21.
[2]. Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096.
[3]. Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–78.
[4]. Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–5.
[5]. Li X, Han J, Yang J, Zhang H. The structural biology of type III CRISPR-Cas systems. J Struct Biol. 2024;216:108070.
[6]. Liu Z, Shi M, Ren Y, Xu H, Weng S, Ning W, et al. Recent advances and applications of CRISPR-Cas9 in cancer immunotherapy. Mol Cancer. 2023;22:35.
[7]. Ferrando J, Filluelo O, Zeigler DR, Picart P. Barriers to simultaneous multilocus integration in Bacillus subtilis tumble down: development of a straightforward screening method for the colorimetric detection of one-step multiple gene insertion using the CRISPR-Cas9 system. Microb Cell Fact. 2023;22:21.
[8]. Jiang F, Doudna JA. CRISPR-Cas9 Structures and Mechanisms. Annu Rev Biophys. 2017;46:505–29.
[9]. Goto T, Yogo K, Hochi S, Hirabayashi M. Characterization of homozygous Foxn1 mutations induced in rat embryos by different delivery forms of Cas9 nuclease. Mol Biol Rep. 2023;50:1231–9.
[10]. Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–91.
[11]. Nuñez JK, Chen J, Pommier GC, Cogan JZ, Replogle JM, Adriaens C, et al. Genome-wide programmable transcriptional memory by CRISPR-based epigenome editing. Cell. 2021;184:2503-2519.e17.
[12]. Chuai G, Ma H, Yan J, Chen M, Hong N, Xue D, et al. DeepCRISPR: optimized CRISPR guide RNA design by deep learning. Genome Biology. 2018;19:80.
[13]. Sander JD, Maeder ML, Reyon D, Voytas DF, Joung JK, Dobbs D. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 2010;38:W462-468.
[14]. Heigwer F, Kerr G, Boutros M. E-CRISP: fast CRISPR target site identification. Nat Methods. 2014;11:122–3.
[15]. Dobson L, Reményi I, Tusnády GE. CCTOP: a Consensus Constrained TOPology prediction web server. Nucleic Acids Res. 2015;43:W408-412.
[16]. Zhang X-H, Tee LY, Wang X-G, Huang Q-S, Yang S-H. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Mol Ther Nucleic Acids. 2015;4:e264.
[17]. Rahman MM, Tollefsbol TO. Targeting cancer epigenetics with CRISPR-dCAS9: Principles and prospects. Methods. 2021;187:77–91.
[18]. Concordet J-P, Haeussler M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018;46:W242–5.
[19]. Labun K, Montague TG, Krause M, Torres Cleuren YN, Tjeldnes H, Valen E. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019;47:W171–4.
[20]. Manghwar H, Li B, Ding X, Hussain A, Lindsey K, Zhang X, et al. CRISPR/Cas Systems in Genome Editing: Methodologies and Tools for sgRNA Design, Off-Target Evaluation, and Strategies to Mitigate Off-Target Effects. Advanced Science. 2020;7:1902312.
[21]. Singh V. Chapter One - An introduction and use of the CRISPR-Cas systems. In: Singh V, editor. Progress in Molecular Biology and Translational Science [Internet]. Academic Press; 2021 [cited 2024 Jul 18]. p. 1–10. Available from: https://www.sciencedirect.com/science/article/pii/S1877117320301812
[22]. Sharpe JJ, Cooper TA. Unexpected consequences: exon skipping caused by CRISPR-generated mutations. Genome Biol. 2017;18:109.
[23]. Hanna RE, Doench JG. Design and analysis of CRISPR–Cas experiments. Nat Biotechnol. 2020;38:813–23.
[24]. Lin Y-Q, Feng K-K, Lu J-Y, Le J-Q, Li W-L, Zhang B-C, et al. CRISPR/Cas9-based application for cancer therapy: Challenges and solutions for non-viral delivery. Journal of Controlled Release. 2023;361:727–49.
[25]. Bernard BE, Landmann E, Jeker LT, Schumann K. CRISPR/Cas-based Human T cell Engineering: Basic Research and Clinical Application. Immunology Letters. 2022;245:18–28.
[26]. Çerçi B, Uzay IA, Kara MK, Dinçer P. Clinical trials and promising preclinical applications of CRISPR/Cas gene editing. Life Sciences. 2023;312:121204.
[27]. Wang X, Xu G, Johnson WA, Qu Y, Yin D, Ramkissoon N, et al. Long sequence insertion via CRISPR/Cas gene-editing with transposase, recombinase, and integrase. Current Opinion in Biomedical Engineering. 2023;28:100491.
Cite this article
Feng,J. (2024). Instrumentation and application of the CRISPR-Cas system. Theoretical and Natural Science,60,128-133.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 4th International Conference on Biological Engineering and Medical Science
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–21.
[2]. Doudna JA, Charpentier E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096.
[3]. Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–78.
[4]. Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–5.
[5]. Li X, Han J, Yang J, Zhang H. The structural biology of type III CRISPR-Cas systems. J Struct Biol. 2024;216:108070.
[6]. Liu Z, Shi M, Ren Y, Xu H, Weng S, Ning W, et al. Recent advances and applications of CRISPR-Cas9 in cancer immunotherapy. Mol Cancer. 2023;22:35.
[7]. Ferrando J, Filluelo O, Zeigler DR, Picart P. Barriers to simultaneous multilocus integration in Bacillus subtilis tumble down: development of a straightforward screening method for the colorimetric detection of one-step multiple gene insertion using the CRISPR-Cas9 system. Microb Cell Fact. 2023;22:21.
[8]. Jiang F, Doudna JA. CRISPR-Cas9 Structures and Mechanisms. Annu Rev Biophys. 2017;46:505–29.
[9]. Goto T, Yogo K, Hochi S, Hirabayashi M. Characterization of homozygous Foxn1 mutations induced in rat embryos by different delivery forms of Cas9 nuclease. Mol Biol Rep. 2023;50:1231–9.
[10]. Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol. 2016;34:184–91.
[11]. Nuñez JK, Chen J, Pommier GC, Cogan JZ, Replogle JM, Adriaens C, et al. Genome-wide programmable transcriptional memory by CRISPR-based epigenome editing. Cell. 2021;184:2503-2519.e17.
[12]. Chuai G, Ma H, Yan J, Chen M, Hong N, Xue D, et al. DeepCRISPR: optimized CRISPR guide RNA design by deep learning. Genome Biology. 2018;19:80.
[13]. Sander JD, Maeder ML, Reyon D, Voytas DF, Joung JK, Dobbs D. ZiFiT (Zinc Finger Targeter): an updated zinc finger engineering tool. Nucleic Acids Res. 2010;38:W462-468.
[14]. Heigwer F, Kerr G, Boutros M. E-CRISP: fast CRISPR target site identification. Nat Methods. 2014;11:122–3.
[15]. Dobson L, Reményi I, Tusnády GE. CCTOP: a Consensus Constrained TOPology prediction web server. Nucleic Acids Res. 2015;43:W408-412.
[16]. Zhang X-H, Tee LY, Wang X-G, Huang Q-S, Yang S-H. Off-target Effects in CRISPR/Cas9-mediated Genome Engineering. Mol Ther Nucleic Acids. 2015;4:e264.
[17]. Rahman MM, Tollefsbol TO. Targeting cancer epigenetics with CRISPR-dCAS9: Principles and prospects. Methods. 2021;187:77–91.
[18]. Concordet J-P, Haeussler M. CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 2018;46:W242–5.
[19]. Labun K, Montague TG, Krause M, Torres Cleuren YN, Tjeldnes H, Valen E. CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019;47:W171–4.
[20]. Manghwar H, Li B, Ding X, Hussain A, Lindsey K, Zhang X, et al. CRISPR/Cas Systems in Genome Editing: Methodologies and Tools for sgRNA Design, Off-Target Evaluation, and Strategies to Mitigate Off-Target Effects. Advanced Science. 2020;7:1902312.
[21]. Singh V. Chapter One - An introduction and use of the CRISPR-Cas systems. In: Singh V, editor. Progress in Molecular Biology and Translational Science [Internet]. Academic Press; 2021 [cited 2024 Jul 18]. p. 1–10. Available from: https://www.sciencedirect.com/science/article/pii/S1877117320301812
[22]. Sharpe JJ, Cooper TA. Unexpected consequences: exon skipping caused by CRISPR-generated mutations. Genome Biol. 2017;18:109.
[23]. Hanna RE, Doench JG. Design and analysis of CRISPR–Cas experiments. Nat Biotechnol. 2020;38:813–23.
[24]. Lin Y-Q, Feng K-K, Lu J-Y, Le J-Q, Li W-L, Zhang B-C, et al. CRISPR/Cas9-based application for cancer therapy: Challenges and solutions for non-viral delivery. Journal of Controlled Release. 2023;361:727–49.
[25]. Bernard BE, Landmann E, Jeker LT, Schumann K. CRISPR/Cas-based Human T cell Engineering: Basic Research and Clinical Application. Immunology Letters. 2022;245:18–28.
[26]. Çerçi B, Uzay IA, Kara MK, Dinçer P. Clinical trials and promising preclinical applications of CRISPR/Cas gene editing. Life Sciences. 2023;312:121204.
[27]. Wang X, Xu G, Johnson WA, Qu Y, Yin D, Ramkissoon N, et al. Long sequence insertion via CRISPR/Cas gene-editing with transposase, recombinase, and integrase. Current Opinion in Biomedical Engineering. 2023;28:100491.









