1. Introduction
Weight control is a significant problem among teenagers. One popular method used to control weight is eating less, also known as starvation. However, this can have detrimental effects on multiple aspects of human health, including an increased risk of death from cardiovascular disease [1]. The impact of starvation on metabolic processes and associated molecular mechanisms has been extensively studied [2][3]. In the meantime, adolescents experience rapid growth and development, which requires a lot of energy for neuronal development [4][5]. Therefore, long-term starvation may affect their ability to learn and remember, but this remains untested.
In this study, we investigated this issue using Caenorhabditis elegans (C. elegans), a commonly used animal model in learning and memory research [6]. This model has several advantages, including a short life cycle, a fully sequenced genome, a well-mapped neural system, the ease of obtainment and maintenance, as well as the ability to self-fertilize [6].
In C. elegans, the main types of learning behaviours are associative and non-associative learning [7]. Non-associative learning refers to a change in response strength to a single stimulus [8], while associative learning involves making an association between two stimuli, using one as a cue to avoid an adverse environment or find food, such as in the salt attractive assay associated with starvation [6]. Previous research has shown that the duration of starvation is an important factor in the impairment of learning ability. For example, starving C. elegans for 24 hours significantly reduced their learning ability by impairing dendrite morphology [9], while starving for 1-2 hours affected benzaldehyde chemotaxis, but 30 minutes of starvation did not have the same effect [10]. However, it is still unclear whether starvation treatment has a continuous effect on learning ability and whether longer durations have a more severe impact on learning and memory. Therefore, in this study, we included durations of 30 minutes, 1 hour, 2 hours, 6 hours, and 12 hours to test the effect of starvation on associative and non-associative learning, as well as some essential vital functions of C. elegans [11].
Genetics has also been recognised as a major regulator of learning behaviours [12]. To determine if the genetics of learning and memory are affected by starvation during development, we performed RT-qPCR to monitor the expression of learning and memory-related genes, including those related to the glutamate system (eat-4/glr-1/nmr-1/nmr-2/casy-1), serotonin system (tph-1), and dopamine system (cat-2/dop-1/flp-34) [12].
2. Method
2.1. Questionnaire-based investigation
A questionnaire was conducted to assess the perceptions and practices of Chinese teenagers regarding weight control. Thirty-nine teenagers aged 15-18 were randomly selected from 10 high schools in Shanghai. The questionnaire included questions about gender, willingness to control weight, reasons for weight control, weight control plans and implementation, and any side effects experienced during the weight control period, particularly negative impacts on learning ability.
2.2. Maintenance and synchronization
Figure 1. The L4 C. elegans under microscopy.
C. elegans were cultured according to the method described by Stiernagle [13], with minor modifications. Specifically, the worms were grown on nematode growth medium (NGM) and fed with E. coli strain OP50 at a temperature of 22.5 ℃. The worm strain used was the wild-type Bristol N2 strain provided by the Caenorhabditis Genetic Center. Synchronisation was achieved by selecting larvae at the L4 stage and transferring them to a new plate every 3 days to maintain a population of similar age (Fig. 1).
2.3. Egg-laying assay
Synchronised L4 worms were utilised to investigate the impact of starvation on egg-laying capacity. The groups are outlined in Table 1. The total and average number of eggs laid after 12, 36, and 72 hours of treatment will be calculated. The average number was determined using the following formula:
\( Average=\frac{Total number of eggs}{Total number of worms in each group} \)
2.4. Locomotion assay
Synchronised L4 worms were tested to determine the impact of starvation on their locomotion. The worms were divided into different groups, and their movement was measured by recording videos of them on OP50-free NGM plates. The worms' movements were tracked by capturing images every 500 milliseconds for a total of 10 minutes. This allowed for the calculation of their average movement speed using the following formula:
\( speed=\frac{Total distance moved}{20} \)
2.5. Body size assay
According to the published method [14], synchronised L4 worms were tested for the effect of starvation on body size, specifically the average circumference, in different groups. Images of the worms were captured using a QiBu BSW-CS-5 inverted microscope and a QiBu SZ-512 front microscope, both with a motorised magnification of 40X. The Digital Analysis System was used to analyse all images.
Table 1. The groups in experiments
OP50 Free (Starvation simulation) | Control (5 plates) | |||||
Duration | 30 min | 1 h | 2 h | 6 h | 12 h | |
Sample Size | 40 for each group | |||||
2.6. Plate-tapping assay
Synchronized L4 worms were tested for the impact of starvation on their non-associative learning ability. The worms were individually transferred to new NGM plates, and a gentle tap on the plates was applied in the direction of their heads. This process was repeated until the worms no longer showed any evasion movements. The number of taps required was recorded as the evasive times. [15]
2.7. Chemotaxis assay
To investigate the effect of starvation on non-associative learning, synchronized L4 worms were pre-treated with 5 μl of diacetyl (10 mg/ml) for 5 minutes. After starvation, the worms were transferred to a chemotaxis plate at point O (the origin). Then, 5 μl of diacetyl was dropped at point A and 5 μl of water was dropped at point B. The distance between points A-O and B-O was kept the same (Fig. 2). To immobilize the worms once they reached the different chemicals, 10 μl of NaN3 (1M) was dropped at points A and B. [15]
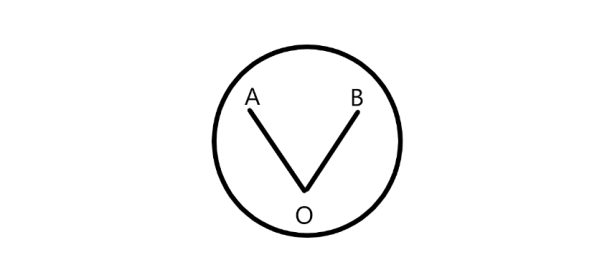
Figure 2. Schematic graph of chemotaxis assay.
2.8. NaCl with copper associative assay
To determine the appropriate duration for worms to stay on copper-containing plates, wild-type L4 worms were treated with 0.5 M copper/Cu and their vitality was observed at 1, 2, and 4 hours. Based on the results, 4 hours was chosen as the test time as it allowed for sufficient exposure to the copper while maintaining worm vitality. The worms were then synchronized to the L4 stage, and any excess E. coli was washed away. They were then subjected to different starvation treatments on 60-mm plates. Each group was further divided into 3 smaller groups, which were placed on standard NGM, NGM with Cu but without NaCl, and NGM with both Cu and 100 mM NaCl, as shown in Table 2. After 4 hours of treatment, the worms were transferred to a specially made chemotaxis plate containing 100 mM NaCl and NaN3 at 20 ℃. The treated worms were placed on the opposite side of the NaCl, and photos were taken after 5 minutes, and number of worms are counted. [16] (Fig. 3)
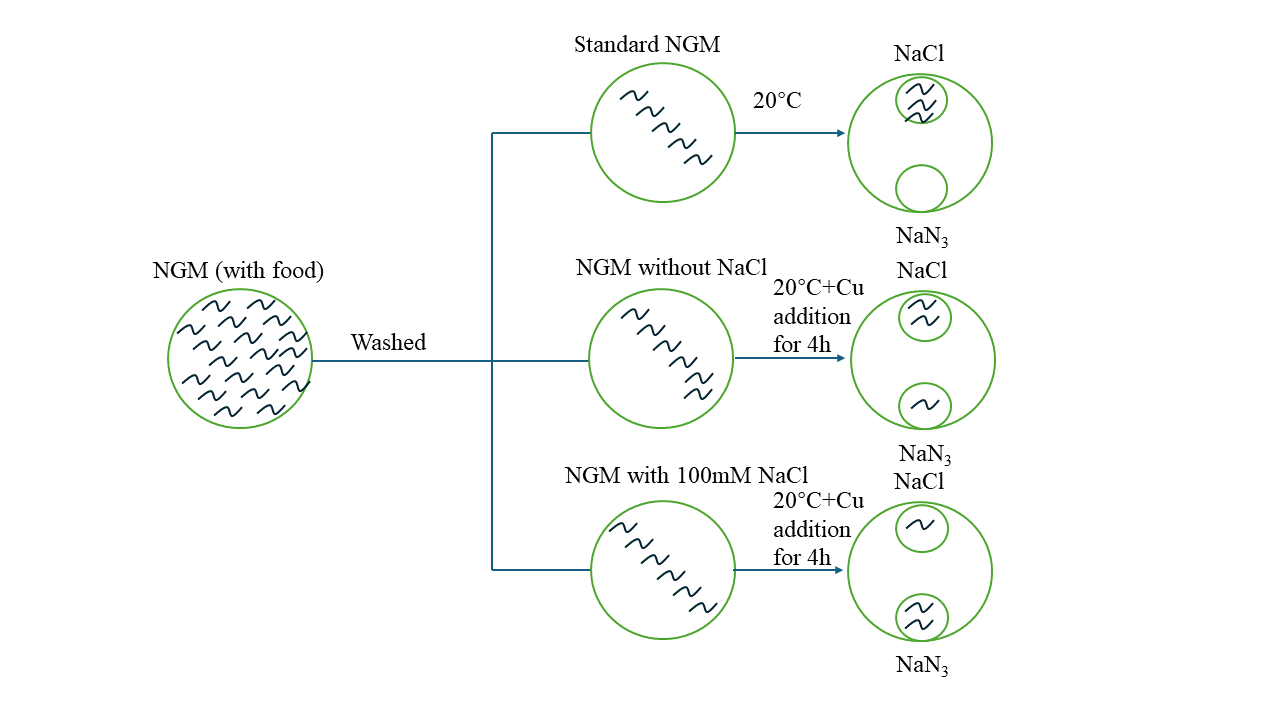
Figure 3. Schematic graph of NaCl with copper associative assay.
Table 2. The experiment and control groups
Experiment Group | Treatment | Sample size |
Standard NGM | 20°C without Cu addition | 100-150 |
NaCl-free NGM | 20°C with 4h Cu addition | |
NGM with 100 mM NaCl | 20°C with 4h Cu addition |
2.9. Construct the phylum tree
The phylogenetic tree for the X gene was created using the Neighbor-joining method in MEGA7, aligning six species (C. elegans, Homo sapiens, Pan troglodytes, Macaca mulatta, Mus musculus, and Xenopus tropicalis). The following parameters were used: Test of Phylogeny-Bootstrap method; No. of Bootstrap Replications-1000; Substitution Type-Nucleotide; Model/Method-Maximum Composite Likelihood; Substitutions to Include-d: Transitions + Transversions; Rates among sites-Uniform rates; Pattern among Lineages-Same (Homogeneous); Gaps/Missing Data Treatment-complete deletion.
2.10. RT-qPCR
Total RNA was extracted from each group [17] for cDNA synthesis. The cDNA was then analyzed using RT-qPCR with tba-1 as the reference gene. The primers were designed using ApE software and can be found in Appendix Table S1.
2.11. Statistical Analysis
Group comparisons were analyzed using Graph Prism version 9.0. The Digital analysis system was used to track and measure the length of each nematode using ImageJ. Student's t-test and one-way ANOVA were used for comparisons between two groups and multiple groups, respectively. Values were expressed as mean ± SEM, and significance was determined at P < 0.05, P < 0.01, and P < 0.001.
3. Results
3.1. Investigation on weight control perceptions and practices found starvation a popular phenomenon among Chinese teenagers
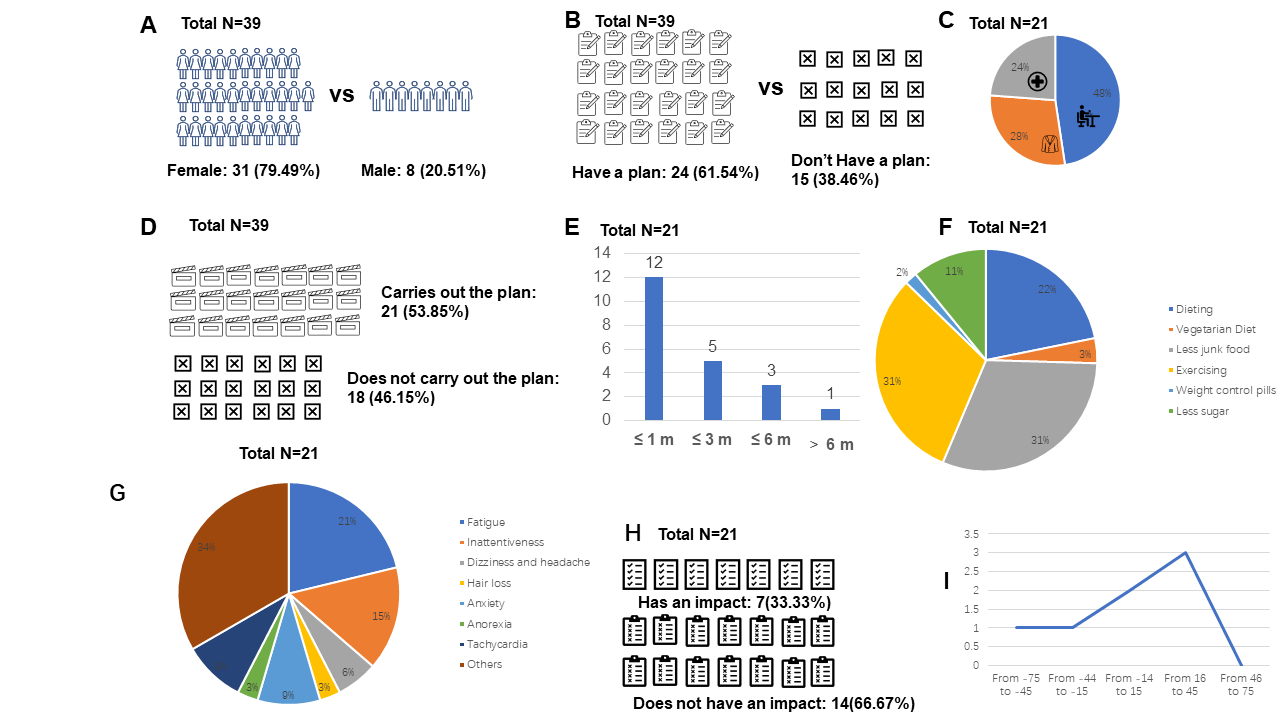
Figure 4. Results of the questionnaire-based research. A: Genders; B: Proportions of those who had a plan; C: Reason for weight control; D: Proportions of those who carried out the plan; E: Duration for weight control; F: Ways of weight control; G: Side effects during weight control; H: The impact of weight control on learning; I: Change in Grades. All data is presented as percentages or averages.
Investigation on weight control perceptions and practices found that starvation is a popular phenomenon among Chinese teenagers. Out of the 39 subjects who responded to the questionnaire, 31 were female and 8 were male, making up 79.49% and 20.51% respectively (Fig. 4A). 61.54% (24/39) of all subjects reported having a plan for weight control (Fig. 4B). Among the reasons given for wanting to control weight (17/39), 52.94% (9/17) cited beauty as their motivation, while the rest mentioned health (Fig. 4C). 53.85% (21/39) of the respondents admitted to carrying out their weight control plan (Fig. 4D). Of those, 55% lasted for 1 month, 25% for 3 months, 15% for half a year, and only 5% lasted for more than half a year (Fig. 4E). The most popular methods of weight control were identified, with diet restriction being one of the top three (Fig. 4F). The reported side effects included inattentiveness and fatigue, each accounting for 21% and 34% respectively (Fig. 4G). Additionally, 33.3% of the respondents stated that starvation had a negative effect on their studies, with an average decrease of 5.71% (Fig. 4H, I).
3.2. Starvation reduced egg laying in C. elegans
Compared to the control group, a 30-minute to 12-hour period of starvation resulted in a significant decrease in the total number of eggs laid from D1 to D3 (P < 0.05). As the duration of starvation increased, there was a continual decrease in the total number of eggs laid over the three days. After 12 hours of starvation, the average number of eggs laid was only 75% of the control group, indicating a significant impact of starvation on egg laying in C. elegans (Fig. 5).
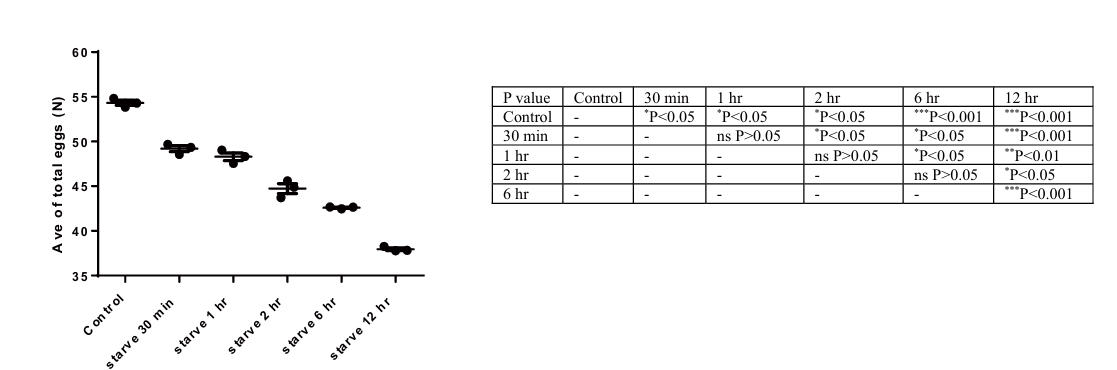
Figure 5. The impact of starvation on egg laying from D1 to D3 of C. elegans. Data are presented as Mean ± SEM. These times independent experiments. * P < 0.05, ** P < 0.01, *** P < 0.001. One-way ANOVA.
The results indicate a significant decrease in egg laying at 12 hours, 36 hours, and 72 hours after starvation treatment compared to the control group (P < 0.05), with the greatest decrease observed at 72 hours (Day 3). This suggests that long-term starvation has a greater impact on egg-laying in C. elegans than short-term starvation (Fig. 6).

Figure 6. The impact of starvation on egg laying at D1, D2 and D3 of C. elegans . Data are presented as Mean ± SEM. These times independent experiments. * P < 0.05, ** P < 0.01, *** P < 0.001. One-way ANOVA.
3.3. Starvation decreased the length of C. elegans
The length of the worm is an important factor in determining its overall health and development. In comparison to the control group, the worms in the starvation group showed a significant decrease in length, particularly in the 12-hour group where the decrease was even more pronounced (p < 0.001 compared to the shorter starvation duration) (Fig. 7).
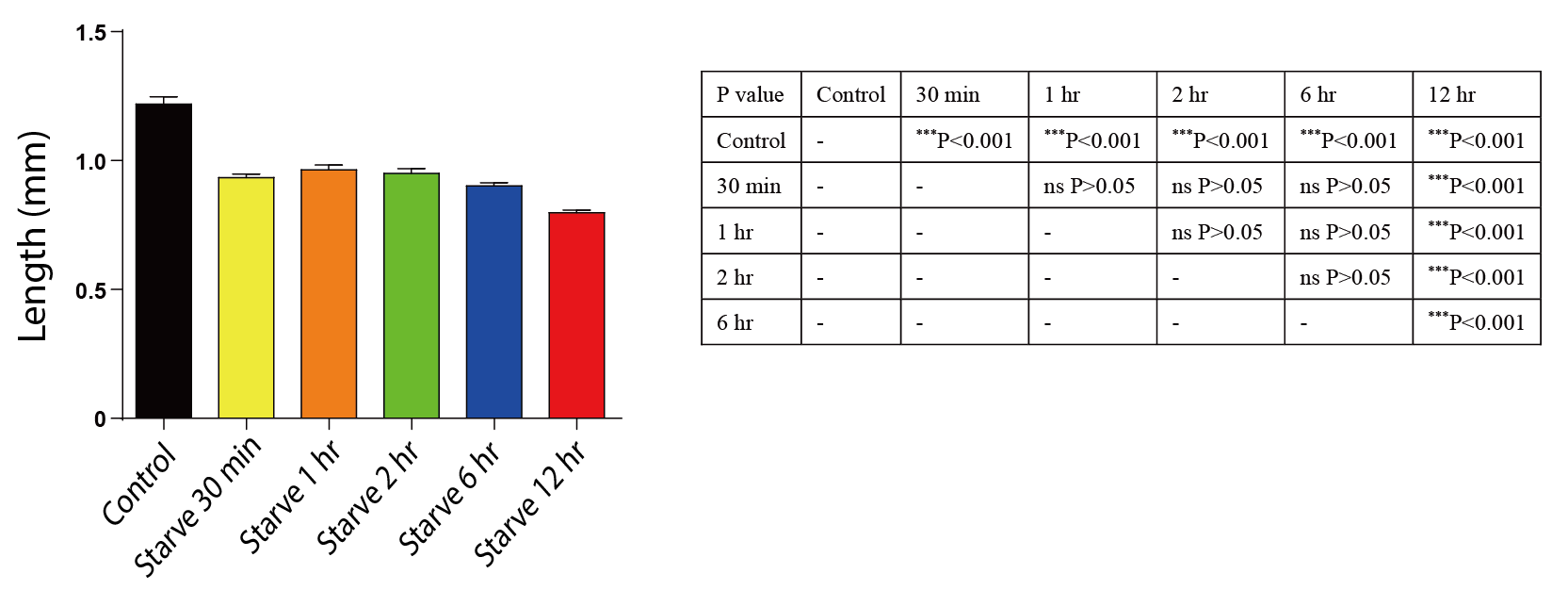
Figure 7. The impact of starvation on the length of C. elegans. Data are presented as Mean ± SEM. * P < 0.05, ** P < 0.01, *** P < 0.001. One-way ANOVA.
3.4. Starvation decreased the locomotion speed of C. elegans
The locomotion speed of C. elegans significantly decreased after being starved for all set durations (p < 0.001). After 30 minutes of starvation, the speed decreased by 6.5%, and after 1 hour it decreased by 37.5%. The decrease continued with longer durations of starvation, with a decrease of 40% after 2 hours, 50% after 6 hours, and 62.5% after 12 hours (Fig. 8). This trend shows that the duration of starvation has a continuous impact on the locomotion speed of C. elegans.
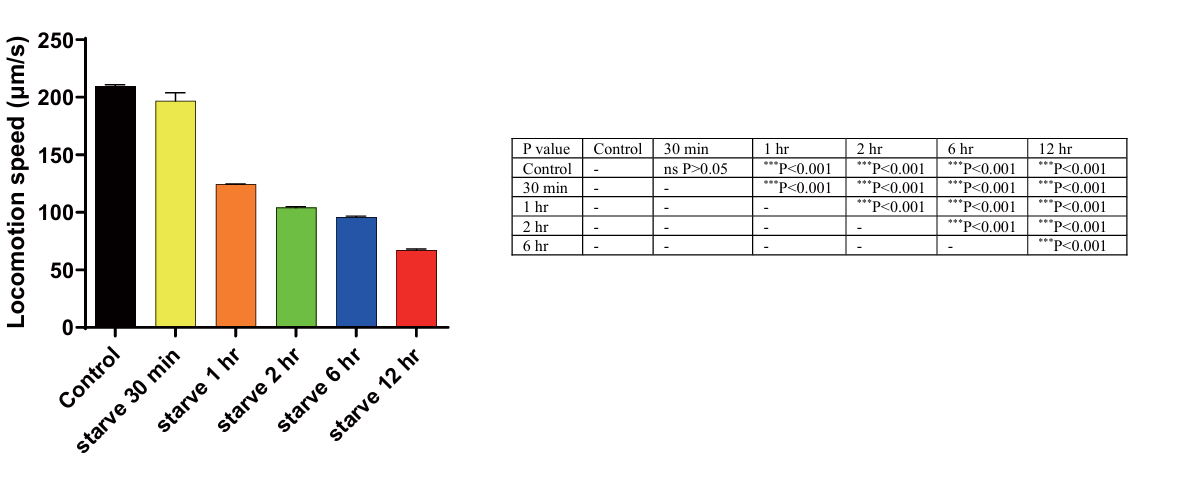
Figure 8. The impact of starvation on the locomotion speed of C. elegans. Data are presented as Mean± SEM. * P < 0.05, **P<0.01, *** P < 0.001. One-way ANOVA.
3.5. Starvation didn’t impair the lifespan of C. elegans significantly
The lifespan assay was performed to identify the impact of short-term starvation on lifespan. As compared to the control group, short-term starvation in 12 hours didn’t have a negative influence on lifespan (P > 0.05) (Fig. 9).
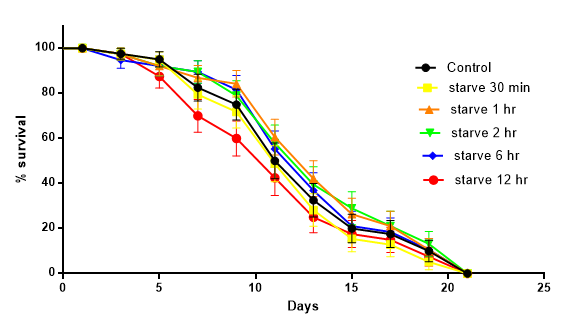
Figure 9. The impact of starvation on lifespan of C. elegans. Data are presented as Mean ± SEM. Logrank-test was used to compare starvation groups with control. P > 0.05.
3.6. Starvation impaired non-associative learning of C. elegans
The effect of starvation on non-associative learning in C. elegans was investigated using the plate-tapping assay and diacetyl chemotaxis assay. Results showed that starvation for 1-12 hours significantly decreased evasion times in the plate-tapping assay (P < 0.001 vs Control) (Fig. 10A). In the diacetyl chemotaxis assay, a decrease in chemotaxis towards diacetyl was observed after 6 hours of starvation (P < 0.05 vs Control). These findings suggest that starvation impairs both mechanosensitive and odor chemotaxis, which are important components of non-associative learning in C. elegans (Fig. 10B).
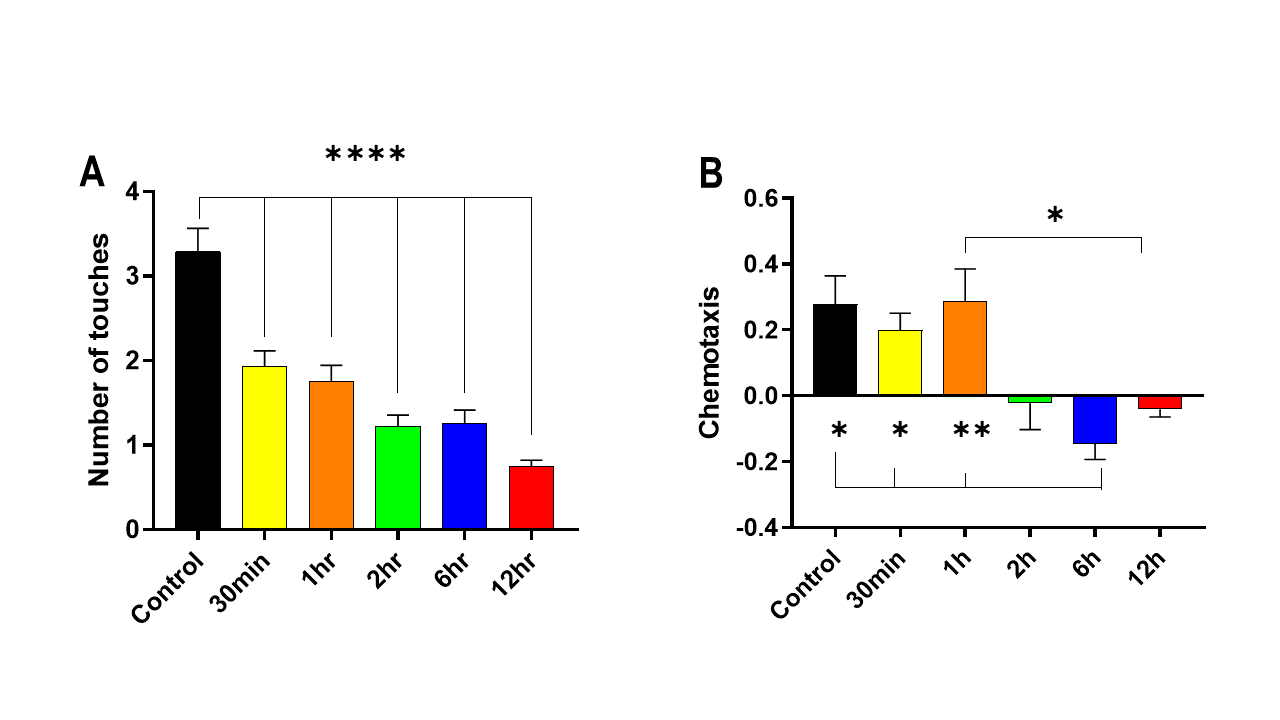
Figure 10. The impact of starvation on non-associative learning of C. elegans. A: Plate-tapping assay. B: Diacytle chemotaxis assay. Data are presented as Mean ± SEM. * P < 0.05, ** P < 0.01, *** P < 0.001. One-way ANOVA.
3.7. Starvation impaired associative learning of C. elegans
The effect of starvation on C. elegans associative learning was identified using the NaCl with copper/Cu associative assay. This assay combines an attractive stimulus, high concentration NaCl, with an aversive stimulus, copper. The results showed that chemotaxis for NaCl did not change significantly on standard NGM plates after starvation (compared to the control group, P > 0.05). This suggests that starvation did not impair the natural chemotaxis of worms toward high-concentration NaCl. However, for the control group that was not starved, co-culture of high-concentration NaCl with copper significantly decreased chemotaxis towards NaCl. This indicates that the non-starved group successfully learned to associate high-concentration NaCl with copper. In contrast, the starvation groups showed a significant decrease in chemotaxis to NaCl after 1, 2, and 6 hours of starvation, but not after 12 hours. This suggests that long-term starvation for 12 hours impaired the ability to associate high-concentration NaCl with copper (Fig. 11).
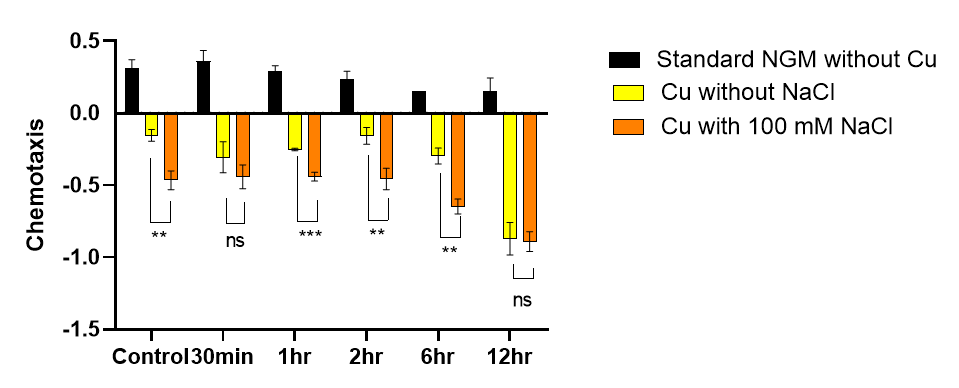
Figure 11. The impact of starvation on NaCl and copper association assay. Data are presented as Mean ± SEM. ** P < 0.01, *** P < 0.001, ns P > 0.05. One-way ANOVA.
3.8. Neurodevelopment-related genes were regulated by starvation in C. elegans
To further investigate the impact of starvation on learning ability in C. elegans, we utilized RT-qPCR to analyze the genetic expressions of key genes involved in the glutamate (eat-4/glr-1/nmr-1/nmr-2/casy-1), serotonin (tph-1), and dopamine (cat-2/dop-1/flp-34) systems. Our results showed a significant increase in relative expression levels of tph-1 and casy-1, as well as a decrease in relative expression level of eat-4 after starvation. Furthermore, the duration of starvation was found to have a direct correlation with the changes in expression levels (Fig. 12).
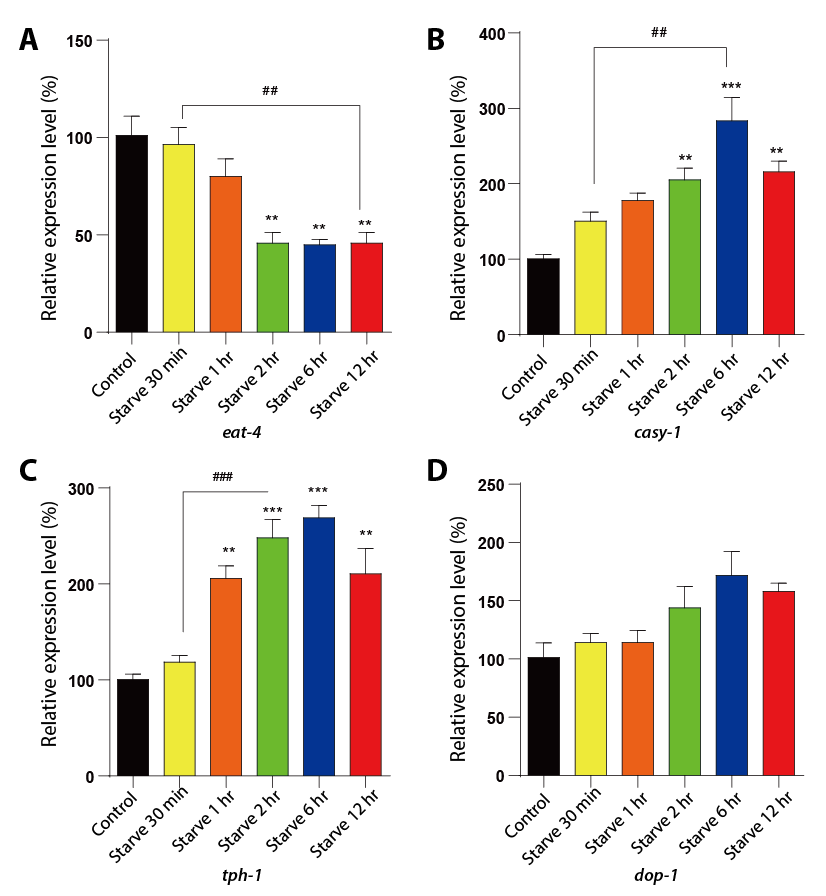
Figure 12. The relative expression level of eat-4, 2022-1, tph-1 and dop-1 genes in C. elegans. There has been an upregulation of the expression of casy-1 and tph-1, with a downregulation of eat-4 expression. A: Relative expression level of eat-4. B: Relative expression level of casy-1. C: Relative expression level of tph-1. D: Relative expression level of dop-1. Data are presented by Mean ± SEM. ** P < 0.01, *** P < 0.001 relative to control group. ## P < 0.01, ### P < 0.001 relative to worms starving 30 min. One-way ANOVA.
3.9. Homology analysis revealed that eat-4 is a homologous gene to human SLC17A7/VGLUT1
To demonstrate the potential for similar mechanisms in other species, we investigated the homologous genes in Homo sapiens, Pan troglodytes, Macaca mulatta, Mus musculus, and Xenopus tropicalis. We chose to focus on eat-4 as it was the only gene that showed down-regulation in response to starvation. Our analysis showed that the C. elegans gene eat-4 is homologous to the human gene SLC17A7/VGLUT1(Fig. 13).
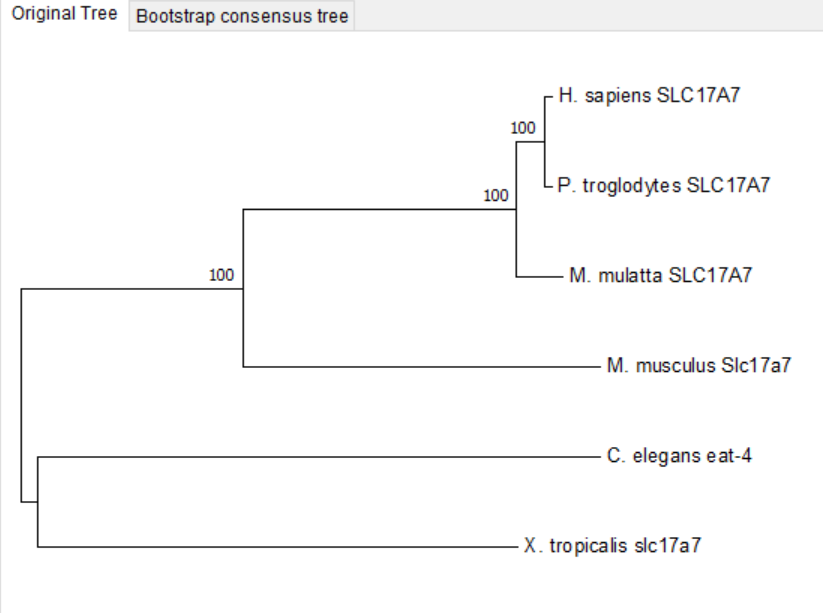
Figure 13. Phylum tree of eat-4 in six species (Homo sapiens, Pan troglodytes, Macaca mulatta, Mus musculus, Caenorhabditis elegans and Xenopus tropicalis.).
4. Discussion
In this study, we conducted questionnaire-based research on Chinese teenagers to investigate the leading method of weight control, which was identified as starvation. We then observed the impact and mechanism of starvation on learning ability in C. elegans. Our results showed a decrease in associative and non-associative learning ability after a period of starvation. To further understand the influence of genetics on starvation, we examined the relative expressions of neurodevelopment-related genes using RT-qPCR. We found that eat-4 was downregulated, while casy-1 and tph-1 were upregulated after starvation. Through homology analysis, we discovered that eat-4 has a homology to the human gene SLC17A7/VGLUT1. Therefore, our study demonstrates that starvation impairs learning ability in C. elegans through the regulation of neurodevelopment-related genes eat-4, casy-1, and tph-1.
4.1. Starvation impairs learning and memory abilities in C. elegans and other species
In this study, we found that starvation had a negative impact on both associative and non-associative learning abilities in C. elegans, particularly when the starvation period lasted for 12 hours. Previous studies on the relationship between starvation and C. elegans have also shown similar results. For example, Guo et al. demonstrated that starvation led to a decrease in backward distance for each tapping movement and an increase in the number of tapping required to habituate to an external cue [9]. Additionally, Campbell et al. found that starvation for over 30 minutes resulted in a decreased copper aversive response, indicating impaired learning abilities [18]. However, there have been conflicting findings, such as Vohra et al. who reported an increase in associative learning ability when the insulin signaling pathway of C. elegans was impaired to mimic starvation [19]. This discrepancy highlights the need for further research on the effects of starvation on learning and memory.
Moreover, there is growing evidence that starvation has a negative impact on learning and memory in various model animals. For instance, Donlea et al. found a significant correlation between extended periods of starvation and decreased learning and memory in D. melanogaster [20]1. In fact, flies that were starved overnight showed impairment in short-term memory and even severe sleep deprivation [20]. Similarly, Briard et al. observed that diet restriction in honeybees affected their ability to associate sensory cues with nectar rewards while foraging, which was regulated by the short neuropeptide F [21]. Overall, these studies suggest a negative correlation between starvation duration and learning and memory abilities.
4.2. The impairment of learning ability by starvation is correlated to the regulation of neurodevelopment-related genes
In this study, starvation caused relatively different regulations in expressions of neurodevelopment-related genes. A 50% downregulation of eat-4, as well as a 100% upregulation of casy-1 and tph-1 upon starvation, were reported in this research.
eat-4 is a learning and memory regulating gene [22], also shown to be necessary for glutamatergic neurotransmission in C. elegans [23], as well as being important in the nocifensive response of C. elegans to obnoxious heat. Mutations in eat-4 in C. elegans led to differences in habituations of to tap-withdrawal effect, specifically having a faster and more complete habituation and slower recovery from habituation [24].
For casy-1, it is involved in learning and memory [25], which helps to explain why its expression becomes increased after more than 2 hours starvation. casy-1 is shown to be a major participant in associative learning [26], and such defects will result in defects in salt chemotaxis learning [27].
tph-1 modulates the feeding behavior in C. elegans, and is also involved in learning and memory [28], where it is mainly characterized to correlate with serotonin signaling, as worms with a deletion in such genes will exhibit a lack of serotonin signaling [29].
Thus, the impact of starvation on the learning and memory abilities in C. elegans is correlated with the upregulation of tph-1 and casy-1, as well as the downregulation of eat-4.
4.3. Longer starving duration impairs the learning ability of C. elegans more severely
Few researchers investigated the relationship between starving duration and learning ability. It demonstrated that after 24 hours of starvation, C. elegans had encountered a reduction in learning ability as reflected by a severely defective PVD dendrite morphology [9], and starvation within 24 hours could prevent the worm from entering Dauer stage [30]. Starvation for 1 hour, 1.5 hours and 2 hours affected C. elegans’s learning ability in benzaldehyde chemotaxis, but starvation for 30 min didn’t cause the effect [10]. Whether starvation treatment will have a continuous effect on learning ability, and whether a longer duration will have a severe effect on learning and memory remains untested. Our study uncovered that generally, prolonged starvation duration was associated with decreased learning and memory abilities. Decreased learning ability for both associative learning and non-associative learning was observed with prolonged starvation duration from 30 minutes to 12 hours.
Thus, a short duration of starvation may result in mild effects on learning and memory, while severe effects can be monitored for a longer duration of starvation.
4.4. The potential impact of starvation on learning ability in human
Questionnaire-based research reported a gap between the number of actual overweight and weight control actions in 13 to 18-year-old American teenagers (11.7% to 43.3%) [31]. Among weight-controlling teenagers, 88.5% of them perceived themselves as overweight, even though the real percentage is only 19.2% [31]. Eating less is one of the top three popular methods used to control weight in teenagers, also known as starvation [32]. In this study, our investigation on Chinese teenagers revealed that 64.86% of the students reported formulating plans to lose weight, and 56.76% had acted, with starvation as one of the high-ranking means (Fig. 1). Almost all subjects reported negative side effects to their bodies at different levels during starvation, like impaired learning ability (Fig. 1).
The popularity of starvation in teenagers has been widely recognized as a social problem, currently, there are limited reports relevant to the problem. The epidemiological research in the investigation suggested that starvation had side effects on multiple areas of performance, with a mostly focused impact on attention deficiency. This may help to explain why some perceived a decrease in score as stated in the results.
Three neurodevelopment-related genes were discovered related to starvation, eat-4, casy-1 and tph-1. Further exploration of the homology of eat-4 in humans was identified by homology analysis. Mutations in eat-4 can result in disruptions of glutamatergic transmission by the positive control of glutaminase activity [33]. The homology in humans, SLC17A7/VGLUT1 [34], transports most of the Glu into the synaptic vesicles [35]. Controlling the activity of VGLUT1 could potentially modulate the efficiency of excitatory neurotransmission and change the filling level of synaptic vesicles [35]. VGLUT1 participates in learning and memory mainly by affecting synaptic Glu transport and long-term potentiation [35]. However, there is little evidence about the impact of starvation on mammalian VGLUT1.
Thus, long-term starvation may impair the learning and memory abilities of teenagers by regulations of neurodevelopment-related genes, which should be further studied in mammals.
5. Conclusion
In this study, we found the popularity of starvation in weight-controlling teenagers in Shanghai, China. Through various assays on essential physiologic indicators as well as learning ability in C. elegans in developing age, we demonstrated a significant correlation between starvation and the impairment of learning ability. This may be attributed to regulations of neurodevelopment-related genes, like eat-4, casy-1 and tph-1. A significant up-regulation of casy-1 and tph-1, as well as a down-regulation of eat-4, has been observed using RT-qPCR. Homology analysis identified the homolog of eat-4 in humans, SLC17A7/VGLUT1, which implied a similar mechanism of starvation in human beings. Therefore, the study demonstrated that starvation could impair the learning ability of C. elegans by regulations of neurodevelopment-related genes.
References
[1]. Friars, D., Walsh, O., & McNicholas, F. (2023). Assessment and management of cardiovascular complications in eating disorders. Journal of Eating Disorders, 11(1). https://doi.org/10.1186/s40337-022-00724-5
[2]. Gottesman, S., & Maurizi, M. R. (2001). CELL BIOLOGY: enhanced: Surviving starvation. Science, 293(5530), 614–615. https://doi.org/10.1126/science.1063371
[3]. Silver, N. (2024, January 19). How long can you live without food? Healthline. https://www.healthline.com/health/food-nutrition/how-long-can-you-live-without-food#water-intake
[4]. Whyte, H., & Findlay, S. (2004). Dieting in adolescence. Paediatrics and Child Health, 9(7), 487–491. https://doi.org/10.1093/pch/9.7.487
[5]. Suryawan, A., Jalaludin, M. Y., Poh, B. K., Sanusi, R. A., Tan, V., Geurts, J. M., & Muhardi, L. (2021). Malnutrition in early life and its neurodevelopmental and cognitive consequences: a scoping review. Nutrition Research Reviews, 35(1), 136–149. https://doi.org/10.1017/s0954422421000159
[6]. Corsi, A. K., Wightman, B., & Chalfie, M. (2015). A Transparent window into biology: A primer on Caenorhabditis elegans. Wormbook, 1–31. https://doi.org/10.1895/wormbook.1.177.1
[7]. Lau, H. L., Timbers, T., Mahmoud, R., & Rankin, C. H. (2012). Genetic dissection of memory for associative and non‐associative learning in Caenorhabditis elegans. Genes, Brain and Behavior, 12(2), 210–223. https://doi.org/10.1111/j.1601-183x.2012.00863.x
[8]. Adachi, T., Kunitomo, H., Tomioka, M., Ohno, H., Okochi, Y., Mori, I., & Iino, Y. (2010). Reversal of Salt Preference Is Directed by the Insulin/PI3K and Gq/PKC Signaling in Caenorhabditis elegans. Genetics, 186(4), 1309–1319. https://doi.org/10.1534/genetics.110.119768
[9]. Guo, N., Wang, J., & Wang, X. (2019). Effect of starvation and high-carbohydrate diet on learning ability of Caenorhabditis elegans. Heliyon, 5(3), e01289. https://doi.org/10.1016/j.heliyon.2019.e01289
[10]. Colbert, H. A., & Bargmann, C. I. (1997). Environmental signals modulate olfactory acuity, discrimination, and memory in Caenorhabditis elegans. Learning & Memory, 4(2), 179–191. https://doi.org/10.1101/lm.4.2.179
[11]. Ishita, Y., Chihara, T., & Okumura, M. (2020). Serotonergic modulation of feeding behavior in Caenorhabditis elegans and other related nematodes. Neuroscience Research, 154, 9–19. https://doi.org/10.1016/j.neures.2019.04.006
[12]. Rahmani, A., & Chew, Y. L. (2021b). Investigating the molecular mechanisms of learning and memory using Caenorhabditis elegans. Journal of Neurochemistry, 159(3), 417–451. https://doi.org/10.1111/jnc.15510
[13]. Stiernagle, T. (2006). Maintenance of C. elegans. Wormbook. https://doi.org/10.1895/wormbook.1.101.1
[14]. Hibshman, J. D., Leuthner, T. C., Shoben, C., Mello, D. F., Sherwood, D. R., Meyer, J. N., & Baugh, L. R. (2018). Nonselective autophagy reduces mitochondrial content during starvation in Caenorhabditis elegans. American Journal of Physiology-cell Physiology, 315(6), C781–C792. https://doi.org/10.1152/ajpcell.00109.2018
[15]. Yang, W., Chen, C., Wang, K., Kwok, H. L., Stern, A., Lo, S. J., & Yang, H. (2019). Reflex and habituation behavior of Caenorhabditis elegans assessed by a mechanical vibration system and image analysis. Journal of Neuroscience Methods, 328, 108415. https://doi.org/10.1016/j.jneumeth.2019.108415
[16]. Rahmani, A., & Chew, Y. L. (2021a). Investigating the molecular mechanisms of learning and memory using Caenorhabditis elegans. Journal of Neurochemistry, 159(3), 417–451. https://doi.org/10.1111/jnc.15510
[17]. Artika, I. M., Dewi, Y. P., Nainggolan, I. M., Siregar, J. E., & Antonjaya, U. (2022). Real-Time polymerase chain Reaction: Current techniques, applications, and role in COVID-19 diagnosis. Genes, 13(12), 2387. https://doi.org/10.3390/genes13122387
[18]. Campbell, J. C., Chin-Sang, I. D., & Bendena, W. G. (2017). A <em>Caenorhabditis elegans</em> Nutritional-status Based Copper Aversion Assay. Journal of Visualized Experiments, 125. https://doi.org/10.3791/55939
[19]. Vohra, M., Lemieux, G. A., Lin, L., & Ashrafi, K. (2017). The beneficial effects of dietary restriction on learning are distinct from its effects on longevity and mediated by depletion of a neuroinhibitory metabolite. PLoS Biology, 15(8), e2002032. https://doi.org/10.1371/journal.pbio.2002032
[20]. Donlea, J. M., Leahy, A., Thimgan, M. S., Suzuki, Y., Hughson, B. N., Sokolowski, M. B., & Shaw, P. J. (2012). foraging alters resilience/vulnerability to sleep disruption and starvation in Drosophila. Proceedings of the National Academy of Sciences of the United States of America, 109(7), 2613–2618. https://doi.org/10.1073/pnas.1112623109
[21]. Briard, E., Carcaud, J., Sandoz, J., Velarde, R., Giurfa, M., & De Brito Sanchez, M. G. (2022). The short neuropeptide F (sNPF) promotes the formation of appetitive visual memories in honey bees. Biology Letters, 18(2). https://doi.org/10.1098/rsbl.2021.0520
[22]. Eat-4 Major facilitator superfamily (MFS) profile domain-containing protein;putative vesicular glutamate transporter eat-4 [Caenorhabditis elegans] - Gene - NCBI. (n.d.). https://www.ncbi.nlm.nih.gov/gene/176291
[23]. Lee, R. Y. N., Sawin, E. R., Chalfie, M., Horvitz, H. R., & Avery, L. (1999). EAT-4, a Homolog of a Mammalian Sodium-Dependent Inorganic Phosphate Cotransporter, Is Necessary for Glutamatergic Neurotransmission inCaenorhabditis elegans. ˜the œJournal of Neuroscience/˜the œJournal of Neuroscience, 19(1), 159–167. https://doi.org/10.1523/jneurosci.19-01-00159.1999
[24]. Wicks, S. R., Roehrig, C. J., & Rankin, C. H. (1996). A dynamic network simulation of the nematode TaP withdrawal Circuit: Predictions concerning synaptic function using behavioral criteria. ˜the œJournal of Neuroscience/˜the œJournal of Neuroscience, 16(12), 4017–4031. https://doi.org/10.1523/jneurosci.16-12-04017.1996
[25]. Casy-1 Calsyntenin C-terminal domain-containing protein;Secreted calsyntenin-1 [Caenorhabditis elegans] - Gene - NCBI. (n.d.). https://www.ncbi.nlm.nih.gov/gene/174006
[26]. Hoerndli, F. J., Walser, M., Hoier, E. F., De Quervain, D., Papassotiropoulos, A., & Hajnal, A. (2009). A Conserved Function of C. elegans CASY-1 Calsyntenin in Associative Learning. PloS One, 4(3), e4880. https://doi.org/10.1371/journal.pone.0004880
[27]. Ikeda, D. D., Duan, Y., Matsuki, M., Kunitomo, H., Hutter, H., Hedgecock, E. M., & Iino, Y. (2008). CASY-1, an ortholog of calsyntenins/alcadeins, is essential for learning in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America, 105(13), 5260–5265. https://doi.org/10.1073/pnas.0711894105
[28]. Tph-1 Biopterin-dependent aromatic amino acid hydroxylase family profile domain-containing protein [Caenorhabditis elegans] - Gene - NCBI. (n.d.). https://www.ncbi.nlm.nih.gov/gene/174227
[29]. Song, B., & Avery, L. (2012). Serotonin Activates Overall Feeding by Activating Two Separate Neural Pathways inCaenorhabditis elegans. ˜the œJournal of Neuroscience/˜the œJournal of Neuroscience, 32(6), 1920–1931. https://doi.org/10.1523/jneurosci.2064-11.2012
[30]. Baugh, L. R., & Hu, P. J. (2020). Starvation responses throughout the Caenorhabditis elegans life cycle. Genetics, 216(4), 837-878.
[31]. Bhurtun, D. D., & Jeewon, R. (2013). Body Weight Perception and Weight Control Practices among Teenagers. ISRN Nutrition, 2013, 1–6. https://doi.org/10.5402/2013/395125
[32]. McDow, K. B., Nguyen, D. T., Herrick, K. A., & Akinbami, L. J. (2019, July 1). Attempts to lose weight among adolescents aged 16–19 in the United States, 2013–2016. https://stacks.cdc.gov/view/cdc/79992
[33]. Rankin, C. H., & Wicks, S. R. (2000). Mutations of theCaenorhabditis elegansBrain-Specific Inorganic Phosphate Transportereat-4Affect Habituation of the Tap–Withdrawal Response without Affecting the Response Itself. ˜the œJournal of Neuroscience/˜the œJournal of Neuroscience, 20(11), 4337–4344. https://doi.org/10.1523/jneurosci.20-11-04337.2000
[34]. Jin, S., Campbell, E. J., Ip, C. K., Layfield, S., Bathgate, R. a. D., Herzog, H., & Lawrence, A. J. (2023). Molecular profiling of VGLUT1 AND VGLUT2 ventral subiculum to nucleus accumbens Shell projections. Neurochemical Research, 48(8), 2490–2501. https://doi.org/10.1007/s11064-023-03921-z
[35]. Du, X., Li, J., Li, M., Yang, X., Qi, Z., Xu, B., Liu, W., Xu, Z., & Deng, Y. (2020). Research progress on the role of type I vesicular glutamate transporter (VGLUT1) in nervous system diseases. Cell & Bioscience, 10(1). https://doi.org/10.1186/s13578-020-00393-4
Cite this article
Mei,S. (2024). Starvation impairs learning ability during development by regulating neurodevelopment-related genes in Caenorhabditis elegans. Theoretical and Natural Science,67,70-83.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 4th International Conference on Biological Engineering and Medical Science
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Friars, D., Walsh, O., & McNicholas, F. (2023). Assessment and management of cardiovascular complications in eating disorders. Journal of Eating Disorders, 11(1). https://doi.org/10.1186/s40337-022-00724-5
[2]. Gottesman, S., & Maurizi, M. R. (2001). CELL BIOLOGY: enhanced: Surviving starvation. Science, 293(5530), 614–615. https://doi.org/10.1126/science.1063371
[3]. Silver, N. (2024, January 19). How long can you live without food? Healthline. https://www.healthline.com/health/food-nutrition/how-long-can-you-live-without-food#water-intake
[4]. Whyte, H., & Findlay, S. (2004). Dieting in adolescence. Paediatrics and Child Health, 9(7), 487–491. https://doi.org/10.1093/pch/9.7.487
[5]. Suryawan, A., Jalaludin, M. Y., Poh, B. K., Sanusi, R. A., Tan, V., Geurts, J. M., & Muhardi, L. (2021). Malnutrition in early life and its neurodevelopmental and cognitive consequences: a scoping review. Nutrition Research Reviews, 35(1), 136–149. https://doi.org/10.1017/s0954422421000159
[6]. Corsi, A. K., Wightman, B., & Chalfie, M. (2015). A Transparent window into biology: A primer on Caenorhabditis elegans. Wormbook, 1–31. https://doi.org/10.1895/wormbook.1.177.1
[7]. Lau, H. L., Timbers, T., Mahmoud, R., & Rankin, C. H. (2012). Genetic dissection of memory for associative and non‐associative learning in Caenorhabditis elegans. Genes, Brain and Behavior, 12(2), 210–223. https://doi.org/10.1111/j.1601-183x.2012.00863.x
[8]. Adachi, T., Kunitomo, H., Tomioka, M., Ohno, H., Okochi, Y., Mori, I., & Iino, Y. (2010). Reversal of Salt Preference Is Directed by the Insulin/PI3K and Gq/PKC Signaling in Caenorhabditis elegans. Genetics, 186(4), 1309–1319. https://doi.org/10.1534/genetics.110.119768
[9]. Guo, N., Wang, J., & Wang, X. (2019). Effect of starvation and high-carbohydrate diet on learning ability of Caenorhabditis elegans. Heliyon, 5(3), e01289. https://doi.org/10.1016/j.heliyon.2019.e01289
[10]. Colbert, H. A., & Bargmann, C. I. (1997). Environmental signals modulate olfactory acuity, discrimination, and memory in Caenorhabditis elegans. Learning & Memory, 4(2), 179–191. https://doi.org/10.1101/lm.4.2.179
[11]. Ishita, Y., Chihara, T., & Okumura, M. (2020). Serotonergic modulation of feeding behavior in Caenorhabditis elegans and other related nematodes. Neuroscience Research, 154, 9–19. https://doi.org/10.1016/j.neures.2019.04.006
[12]. Rahmani, A., & Chew, Y. L. (2021b). Investigating the molecular mechanisms of learning and memory using Caenorhabditis elegans. Journal of Neurochemistry, 159(3), 417–451. https://doi.org/10.1111/jnc.15510
[13]. Stiernagle, T. (2006). Maintenance of C. elegans. Wormbook. https://doi.org/10.1895/wormbook.1.101.1
[14]. Hibshman, J. D., Leuthner, T. C., Shoben, C., Mello, D. F., Sherwood, D. R., Meyer, J. N., & Baugh, L. R. (2018). Nonselective autophagy reduces mitochondrial content during starvation in Caenorhabditis elegans. American Journal of Physiology-cell Physiology, 315(6), C781–C792. https://doi.org/10.1152/ajpcell.00109.2018
[15]. Yang, W., Chen, C., Wang, K., Kwok, H. L., Stern, A., Lo, S. J., & Yang, H. (2019). Reflex and habituation behavior of Caenorhabditis elegans assessed by a mechanical vibration system and image analysis. Journal of Neuroscience Methods, 328, 108415. https://doi.org/10.1016/j.jneumeth.2019.108415
[16]. Rahmani, A., & Chew, Y. L. (2021a). Investigating the molecular mechanisms of learning and memory using Caenorhabditis elegans. Journal of Neurochemistry, 159(3), 417–451. https://doi.org/10.1111/jnc.15510
[17]. Artika, I. M., Dewi, Y. P., Nainggolan, I. M., Siregar, J. E., & Antonjaya, U. (2022). Real-Time polymerase chain Reaction: Current techniques, applications, and role in COVID-19 diagnosis. Genes, 13(12), 2387. https://doi.org/10.3390/genes13122387
[18]. Campbell, J. C., Chin-Sang, I. D., & Bendena, W. G. (2017). A <em>Caenorhabditis elegans</em> Nutritional-status Based Copper Aversion Assay. Journal of Visualized Experiments, 125. https://doi.org/10.3791/55939
[19]. Vohra, M., Lemieux, G. A., Lin, L., & Ashrafi, K. (2017). The beneficial effects of dietary restriction on learning are distinct from its effects on longevity and mediated by depletion of a neuroinhibitory metabolite. PLoS Biology, 15(8), e2002032. https://doi.org/10.1371/journal.pbio.2002032
[20]. Donlea, J. M., Leahy, A., Thimgan, M. S., Suzuki, Y., Hughson, B. N., Sokolowski, M. B., & Shaw, P. J. (2012). foraging alters resilience/vulnerability to sleep disruption and starvation in Drosophila. Proceedings of the National Academy of Sciences of the United States of America, 109(7), 2613–2618. https://doi.org/10.1073/pnas.1112623109
[21]. Briard, E., Carcaud, J., Sandoz, J., Velarde, R., Giurfa, M., & De Brito Sanchez, M. G. (2022). The short neuropeptide F (sNPF) promotes the formation of appetitive visual memories in honey bees. Biology Letters, 18(2). https://doi.org/10.1098/rsbl.2021.0520
[22]. Eat-4 Major facilitator superfamily (MFS) profile domain-containing protein;putative vesicular glutamate transporter eat-4 [Caenorhabditis elegans] - Gene - NCBI. (n.d.). https://www.ncbi.nlm.nih.gov/gene/176291
[23]. Lee, R. Y. N., Sawin, E. R., Chalfie, M., Horvitz, H. R., & Avery, L. (1999). EAT-4, a Homolog of a Mammalian Sodium-Dependent Inorganic Phosphate Cotransporter, Is Necessary for Glutamatergic Neurotransmission inCaenorhabditis elegans. ˜the œJournal of Neuroscience/˜the œJournal of Neuroscience, 19(1), 159–167. https://doi.org/10.1523/jneurosci.19-01-00159.1999
[24]. Wicks, S. R., Roehrig, C. J., & Rankin, C. H. (1996). A dynamic network simulation of the nematode TaP withdrawal Circuit: Predictions concerning synaptic function using behavioral criteria. ˜the œJournal of Neuroscience/˜the œJournal of Neuroscience, 16(12), 4017–4031. https://doi.org/10.1523/jneurosci.16-12-04017.1996
[25]. Casy-1 Calsyntenin C-terminal domain-containing protein;Secreted calsyntenin-1 [Caenorhabditis elegans] - Gene - NCBI. (n.d.). https://www.ncbi.nlm.nih.gov/gene/174006
[26]. Hoerndli, F. J., Walser, M., Hoier, E. F., De Quervain, D., Papassotiropoulos, A., & Hajnal, A. (2009). A Conserved Function of C. elegans CASY-1 Calsyntenin in Associative Learning. PloS One, 4(3), e4880. https://doi.org/10.1371/journal.pone.0004880
[27]. Ikeda, D. D., Duan, Y., Matsuki, M., Kunitomo, H., Hutter, H., Hedgecock, E. M., & Iino, Y. (2008). CASY-1, an ortholog of calsyntenins/alcadeins, is essential for learning in Caenorhabditis elegans. Proceedings of the National Academy of Sciences of the United States of America, 105(13), 5260–5265. https://doi.org/10.1073/pnas.0711894105
[28]. Tph-1 Biopterin-dependent aromatic amino acid hydroxylase family profile domain-containing protein [Caenorhabditis elegans] - Gene - NCBI. (n.d.). https://www.ncbi.nlm.nih.gov/gene/174227
[29]. Song, B., & Avery, L. (2012). Serotonin Activates Overall Feeding by Activating Two Separate Neural Pathways inCaenorhabditis elegans. ˜the œJournal of Neuroscience/˜the œJournal of Neuroscience, 32(6), 1920–1931. https://doi.org/10.1523/jneurosci.2064-11.2012
[30]. Baugh, L. R., & Hu, P. J. (2020). Starvation responses throughout the Caenorhabditis elegans life cycle. Genetics, 216(4), 837-878.
[31]. Bhurtun, D. D., & Jeewon, R. (2013). Body Weight Perception and Weight Control Practices among Teenagers. ISRN Nutrition, 2013, 1–6. https://doi.org/10.5402/2013/395125
[32]. McDow, K. B., Nguyen, D. T., Herrick, K. A., & Akinbami, L. J. (2019, July 1). Attempts to lose weight among adolescents aged 16–19 in the United States, 2013–2016. https://stacks.cdc.gov/view/cdc/79992
[33]. Rankin, C. H., & Wicks, S. R. (2000). Mutations of theCaenorhabditis elegansBrain-Specific Inorganic Phosphate Transportereat-4Affect Habituation of the Tap–Withdrawal Response without Affecting the Response Itself. ˜the œJournal of Neuroscience/˜the œJournal of Neuroscience, 20(11), 4337–4344. https://doi.org/10.1523/jneurosci.20-11-04337.2000
[34]. Jin, S., Campbell, E. J., Ip, C. K., Layfield, S., Bathgate, R. a. D., Herzog, H., & Lawrence, A. J. (2023). Molecular profiling of VGLUT1 AND VGLUT2 ventral subiculum to nucleus accumbens Shell projections. Neurochemical Research, 48(8), 2490–2501. https://doi.org/10.1007/s11064-023-03921-z
[35]. Du, X., Li, J., Li, M., Yang, X., Qi, Z., Xu, B., Liu, W., Xu, Z., & Deng, Y. (2020). Research progress on the role of type I vesicular glutamate transporter (VGLUT1) in nervous system diseases. Cell & Bioscience, 10(1). https://doi.org/10.1186/s13578-020-00393-4









