1. Introduction
According to statistics in 2020, primary liver cancer has a mortality rate of more than 90%, and is the sixth most commonly diagnosed cancer in the world. In most regions, the incidence of liver cancer in men is generally 2 to 3 times that of women, which may be caused by some bad lifestyle habits Primary liver cancer mainly includes hepatocellular carcinoma (HCC), intrahepatic cholangiocarcinoma and other rare types, while the main causes of HCC are hepatitis B virus (HBV) and hepatitis C virus (HCV), of which HCC accounts for about 75% to 85%, and intrahepatic cholangiocarcinoma accounts for about 10% to 15% [1, 2]. At present, the exploration of therapeutic targets of liver cancer is not perfect, and various mutated genes detected in liver cancer cell models may be studied as target genes for the treatment of liver cancer.
Hepatocyte nuclear factor 4 α (HNF4α) is a transcription factor (TF) enriched in organs such as the liver, gallbladder, pancreas, kidney, etc., related to the formation and development of these organs. HNF4α regulates the cell cycle and life activities of hepatic progenitor cells by regulating TF. Mutations in HNF4A can lead to lesions in related organs. HNF4α dysregulation affects multiple diseases, including Maturity-Onset Diabetes of the Young Type 1 (MODY-1), Glycogen Storage Disease Type I (GSD I), especially in liver cancer [3, 4]. Studies have shown that HNF4A expression is significantly decreased in liver cancer cells.
In this review, the role of HNF4α in liver cancer is discussed. The studies of expression of HNF4A in hepatoma cell models and regulatory pathways are reviewed, providing theoretical basis for the discovery of therapeutic targets for hepatoma.
2. HNF4A gene and HNF4α protein function
2.1. HNF4A gene
HNF4α is a protein encoded by the HNF4A gene on human chromosome 20 and on mouse chromosome 2 [5]. HNF4α is a nuclear receptor. HNF4α is a dimeric transcription factor encoded by two independent promoters, P1 and P2. These two promoters produce 12 transcript samples through alternative cleavage, resulting in a wide variety of isoforms. These isoforms are numbered as HNF4A 1-12, with the first 6 regulated by the P1 promoter and the last 6 regulated by the P2 promoter. These subtypes are expressed differently between tissues. The isoforms are relatively conserved in their functional domains. Thus, general studies often refer to them collectively as "HNF4α" [4, 6].
2.2. HNF4α protein structure and function
The list of authors should be indented 25 mm to match the abstract. HNF4α binds to DNA sequences in the form of a homodimer. HNF4α can also bind to other hepatocellular nuclear factors (HNFs). However, since the amino acid residues of HNF4α inhibit this heterodimeric complex, only HNF4γ containing the same amino acid residues as HNF4α can form heterodimers with HNF4α [5]. Most HNF4A are homodimers and consist of two identical polypeptide chains. There are five domains, A/B Domain, C Domain, D Domain, E/F Domain, and Ligand-Binding Domain (LBD) [7].
The C domain contains the DNA-binding domain (DBD). The DBD contains two zinc fingers that specifically recognize and bind to the HNF4 response element (HNF4 RE) on the DNA sequence, which allows HNF4α to bind precisely and stably to the upstream sequence of the target gene [8,9].
Another key domain on HNF4A is LBD. LBD has a hydrophobic domain, and the conformation and hydrophobicity of this domain can enable HNF4α to bind to specific ligands, thus exhibiting a strong ligand-dependent effect. When a specific ligand is bound, the conformational change of LBD and can co-activate transcription with the LXXLL motif or co-inhibit transcription with the LXXXIXXX(I/L) motif. It has been analyzed that the first amino acid of most LXXLL sequences in RIP-140 is hydrophobic, so it is speculated that the LXXLL sequences interact with each other due to their preference for the hydrophobic domain of LBD [8]. The hinge region of LBD binds to the DNA-binding domain (DBD), where the sequence that binds to LBD is usually highly conserved. After combining with LBD, DBD can change the conformation of LBD to form dimerization, or it can form dimerization on its own in the absence of LBD. The dimerization of DBD in LBD with HNF4α can improve the stability of the DBD-HNF4α complex [4, 8].
3. HNF4α acts as a transcription factor to regulate gene expression
As a transcription factor, HNF4α can bind to the upstream sequence of the target gene and plays a role in regulating gene expression. Therefore, it is important to analyze how HNF4A binds or is recruited to DNA sequences. Studies have shown that HNF4α will be recruited to the target sequence by interacting with the Mixed-Lineage Leukemia 4(MLL4) complex, affecting H3K4me1 near the binding domain of HNF4α. Histone modifications such as H3K27ac and chromosome accessibility affect chromosome openness and regulate downstream gene expression [10]. In addition, these histone modifications also affect the binding of HNF4α to target sequences. Histones change the openness and accessibility of chromatin through methylation, acetylation, ubiquitination, phosphorylation and other ways, thus affecting the binding ability of proteins to DNA sequences. Studies have shown that some signal transduction-dependent specific TF will recruit MLL4 in the upstream enhancer sequence of target genes, resulting in histone modifications such as H3K4me1. These results indicate that the ability of HNF4α to bind to DNA sequence is largely related to histone modification [10].
HNF4α is a member of the nuclear receptor family. It is responsible for regulating the expression of many genes, including TET family genes, glucokinase (GCK), albumin (ALB), fatty acid synthase (FAS), cholesterol 7 alpha-hydroxylase (CYP7A1), Solute Carrier Family 22 Member 4 (SLC22A4), Apolipoprotein E (APOE) and many other genes are involved in regulating a series of physiological processes such as glycolipid metabolism and bile acid metabolism in liver, gallbladder, pancreas, kidney and other organs. In the liver, HNF4α is required for normal growth and development. A study showed that TET proteins (Ten-Eleven Translocation proteins) are a class of enzymes that play an important role in DNA epigenetics, mainly involved in the process of DNA demethylation. TET proteins oxidize 5 mC to 5 hmC during epigenetic processes, thereby regulating the degree of chromatin openness. This regulatory effect involves the regulation of the differentiation ability of liver cells. The histone-modified variant H3K4me1 can mark the enhancer upstream of the TET gene, which is beneficial to increase the degree of chromatin openness, thereby activating the expression of downstream genes. Therefore, HNF4α, as a transcription factor to participate in the metabolism and proliferation of liver cells, requires further research to better elucidate its role in liver cancer [11].
4. HNF4α regulates downstream target gene expression
4.1. Effect of P1/P2 regulated subtypes on HNF4α function
Studies have analyzed the correlation between carcinogenesis pathology and different HNF4A subtypes by culturing different human GC cell lines using short hairpin RNA (shRNA) transfected variants of the HNF4 family into cell lines and cultured them to analyze P1-HNF4A and P2-HNF4A levels using western blot as well as RNA sequencing and qRT-PCR, while also designing experiments to validate the results at the individual mouse level. They found that overexpression of P1-HNF4A led to proliferation and metastasis of human GC cell lines, while overexpression of P2-HNF4A had no effect. However, both P1- and P2-HNF4A led to a significant increase in mRNA levels, which they attributed to the fact that P1-HNF4A was generally higher than P2-HNF4A With stronger trans-activation function and C-C Motif Chemokine Ligand 15 (CCL15) As a chemokine, they detected that P1-HNF4A more significantly activates the expression of the CCL15 gene, which is also involved in carcinogenesis as a direct target of P1-HNF4A [12].
4.2. HNF4α-HIC1 balance shows pathway that HNF4α regulates expression of FUF/FDF
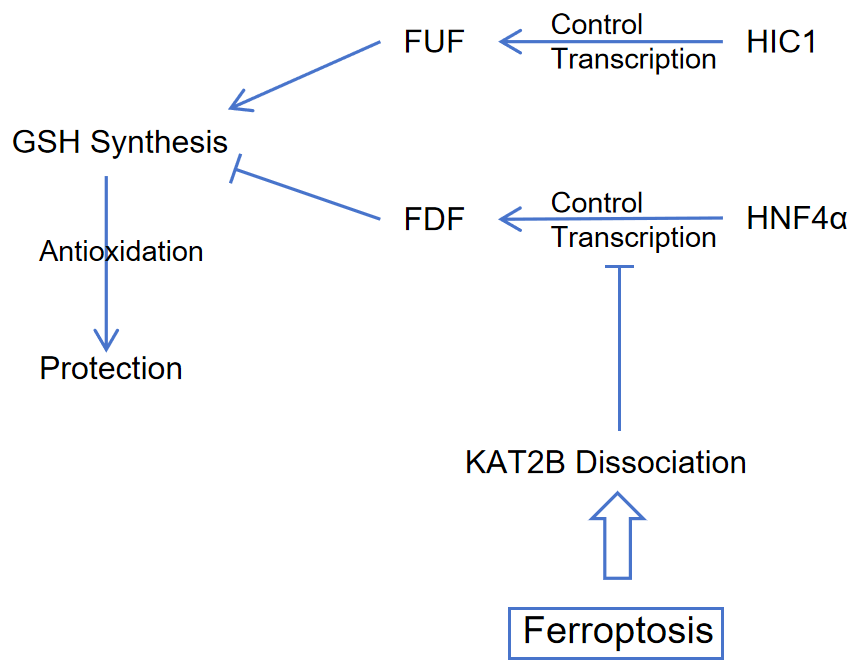
Another example of HNF4A's involvement in the regulatory process of liver cancer formation was presented. Studies were conducted to analyze the regulatory mechanism of HNF4α in the process of liver cancer caused by iron death. They analyzed how the expression of Ferroptosis Upregulator Factor (FUF) and Ferroptosis Downregulator Factor (FDF) by constructing HNF4α/HIC1 deletion and HNF4α/HIC1 overexpression models in mouse models of liver cancer, and designing molecular experiments and real-time sequencing regulated by HNF4α and HIC1. The researchers located a protein in which HNF4A is recruited, KAT2B. When iron death occurs, KAT2B degrades and causes HNF4α to fail to bind to the upstream promoter of FDF, thereby regulating FDF expression. At the same time, they found the antagonistic relationship between HNF4α and HIC1 and studied their clinical significance. HNF4A expression was up-regulated while HIC1 expression was down-regulated in the liver cancer model. Therefore, they speculated that the maintenance of HNF4α-HIC1 balance was of great significance in inhibiting the formation of liver cancer [13].
The graphical abstract of ferroptosis process with regulation of HNF4α/HIC1 is summarized in Figure 1.
|
Figure 1. Mechanism of HNF4α/HIC1 regulating expression of FUF/FDF. With the stimulation of ferroptosis, KAT2B dissociate, which reduces binding of HNF4α to FDF promoter. Cells exert antioxidant effects with a low level of FDF, and a high level of FUF enables cells. Abbreviations: HIC1, hypermethylated in cancer 1; HNF4α, hepatocyte nuclear factor 4 alpha; FUF, ferroptosis up-regulated factors; FDF, ferroptosis down-regulated factors; GSH, glutathione; KAT2B, lysine acetyltransferase 2B; Figure credit: original. |
5. Targeting and regulating HNF4α in liver cancer models
After understanding the mechanism by which HNF4A regulates gene expression as a transcription factor, the researchers designed experiments to verify which pathways regulate the expression of HNF4A and elucidated the specific mechanisms of these pathways. The researchers were able to find target factors that act on the upstream sequences of the HNF4A gene and looked at the expression of HNF4A in models that overexpressed or deleted these factors. Table 1 summarizes the research on mechanisms of various factors that regulate HNF4A expression.
5.1. Epigenetic modifications of HNF4A
A study found that Lysine Demethylase 1A (KDM1A) can regulate HNF4A expression. KDM1A enriched on hepatic transposable elements (TEs), thereby inhibiting the histone modification H3K4me1, resulting in HNF4A epigenetic silencing. Researchers verified that KDM1A regulates HNF4A expression through liver-TEs by targeting liver TEs using the CRISPR activation (CRISPRa) system in a hepatocellular carcinoma cell line model. The researchers also obtained evidence that KDM1A interacted with HNF4α to silence the downstream target gene HNF4A through sequencing and molecular experiments. Since it was found that KDM1A does not have a DNA-binding domain (DBD) and cannot directly bind to DNA sequences, the researchers also found that ZMYM3 has a DNA-binding domain (DBD) as an intermediate. ZMYM3 uses zinc-finger protein to locate and bind to DNA sequence regions that are negatively regulated by KDM1A, and recruits KDM1A. This study revealed that KDM1A is an important target for regulating the expression of the HNF4A gene, which provides an important basis for the study of the structure and function of HNF4A [14].
5.2. Downregulate HNF4A promoter activity
Similarly, two studies have found mutations in the HNF1α POU domain to downregulate HNF4A promoter activity and NRF2-mediated STAT3 activation to silence HNF4A expression through miR-4, respectively. In the case of the former, the researchers found that the expression of HNF1α was strongly correlated with HNF4α, and the mutation of the POU domain in the three domains of HNF1α caused HNF1α to be unable to bind to the promoter region of HNF4A normally, which was reflected in the decrease of transcriptional activity of HNF4A. In the case of the latter, the researchers found that NRF2 signaling was activated due to endoplasmic reticulum (ER) stress due to HCV invasion infection. NRF2 activates STAT3, upregulating miR-24 and resulting in silencing of HNF4A expression. They revealed the protective pathway of the NRF2--STAT3--miR-24--HNF4A regulatory chain in response to HCV infection, and also provided a pathway to edit the ability of HNF4A expression.
In addition, in order to construct a mouse model of specific overexpression (OE) of HNF4A in the liver, the researchers constructed a HNF4A-Myc-pLIVE expression vector (cloned HNF4A-Myc into the pLIVE vector), diluted and injected into mice [15, 16].
Thus, further research can focus on finding a pathway that directly regulates HNF4A gene expression, looking for transcription factors upstream of the HNF4A gene, or looking for molecules that can interact with HNF4α. For example, researchers may choose to observe whether pathways that affect apparent modifications are able to influence HNF4A, especially those that affect H3K4me1 modifications. Because both the expression and executive functions of HNF4A are inseparable from the H3K4me1 modification process.
Table 1. Methods of inhibiting HNF4A expression
Authors and year | Model building | Mechanism | Ref. |
Jing, T et al. 2024 | KDM1A was targeted with a CRISPRa system by constructing lentiviral plasmids in aberrant hepatoma cell lines | KDM1A inhibits the expression of HNF4A in aberrant hepatocellular carcinoma cells by inhibiting H3K4me1 in liver-TEs | [14] |
Haque E et al. 2022 | Knockdown (KD) HNF1A by designing siRNAs, or MD simulations using GROMACS to study HNF1A POU domain mutants (e.g. V295F). | The mutation of the gene segment encoding the POU domain of HNF1A inhibits the expression of HNF4A due to its inability to bind to the upstream promoter of HNF4A gene. | [15] |
Aydin Y et al. 2019 | Culturing liver Huh-7.5 cell line and investigating NRF2 pathway by siRNA transfection. | PERK mediates pro-survival signaling in cells with persistent HCV infection through NRF2-mediated signaling and activation of STAT3. In this process, the expression of HNF4A is inhibited by miR-24 and miR-619. | [16] |
6. Conclusion
As a transcription factor, HNF4α usually acts in the upstream sequence of target genes and plays an important role in regulating gene expression. HNF4α is also an important target for the treatment of liver cancer. HNF4α usually forms a homodimer, which binds to DBD by changing the spatial structure of LBD after binding to a specific ligand and then binds to the DNA sequence to form a stable complex. HNF4α can be recruited onto DNA sequences by a number of intermediates, including MLL4, KAT2B, etc. From multiple studies using hepatocellular carcinoma cell models with HNF4A KO/KD/OE, it has been found that several pathways can regulate HNF4A expression, including the KDM1A pathway, NRF2 pathway, and HNF1α pathway, as well as overexpression by transfection of expression plasmids. Through these research of HNF4α's involvement in the regulation of target genes in liver cancer models, HNF4α exerts an important role in the formation and development of liver cancer. Thus, HNF4α has potential clinical value as an essential target gene for treating liver cancer. For some of these pathways, blocking factors can likely become clinical drugs for the treatment of liver cancer.
References
[1]. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A and Bray F 2021 CA Cancer J. Clin. 71 209-249
[2]. Vogel A, Meyer T, Sapisochin G, Salem R and Saborowski A 2022 Lancet 400 1345-1362
[3]. Kotulkar M, Robarts DR and Apte U 2023 Semin. Liver Dis. 43 234-244
[4]. Radi SH, Vemuri K, Martinez-Lomeli J and Sladek FM 2023 Front Endocrinol. (Lausanne) 14 1226173
[5]. Kotulkar M, Robarts DR and Apte U 2023 Semin Liver Dis. 43 234-244
[6]. Ko HL, Zhuo Z and Ren EC 2019 Cell Rep. 26 2549-2557
[7]. Kumar R 2023 Vitam Horm. 123 399-416
[8]. Heery DM, Kalkhoven E, Hoare S and Parker MG 1997 Nature 387 733-736
[9]. Nadal M, et al. 2017 Nat. Commun. 8 14388
[10]. Thakur A, et al. 2024 Commun. Biol. 7 144
[11]. Hu L, et al. 2015 Nature 527 118-122
[12]. Ni Z, Lu W, Li Q, Han C, Yuan T, Sun N and Shi Y 2021 Cancer Biol. Med. 18 530-546
[13]. Zhang X, et al. 2019 Redox Biol. 24 101211
[14]. Jing T, Wei D, Xu X, Wu C, Yuan L, Huang Y, Liu Y, Jiang Y and Wang B 2024 Nat. Commun. 15 5631
[15]. Haque E, Teeli AS, Winiarczyk D, Taguchi M, Sakuraba S, Kono H, Leszczyński P, Pierzchała M and Taniguchi H 2022 Genes (Basel) 13 413
[16]. Aydin Y, et al. 2019 Cancers (Basel) 11 1407
Cite this article
Zhang,N. (2024). Effect of HNF4α on the Process of Liver Cancer and as a Potential Therapeutic Target. Theoretical and Natural Science,65,101-106.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of ICBioMed 2024 Workshop: Computational Proteomics in Drug Discovery and Development from Medicinal Plants
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A and Bray F 2021 CA Cancer J. Clin. 71 209-249
[2]. Vogel A, Meyer T, Sapisochin G, Salem R and Saborowski A 2022 Lancet 400 1345-1362
[3]. Kotulkar M, Robarts DR and Apte U 2023 Semin. Liver Dis. 43 234-244
[4]. Radi SH, Vemuri K, Martinez-Lomeli J and Sladek FM 2023 Front Endocrinol. (Lausanne) 14 1226173
[5]. Kotulkar M, Robarts DR and Apte U 2023 Semin Liver Dis. 43 234-244
[6]. Ko HL, Zhuo Z and Ren EC 2019 Cell Rep. 26 2549-2557
[7]. Kumar R 2023 Vitam Horm. 123 399-416
[8]. Heery DM, Kalkhoven E, Hoare S and Parker MG 1997 Nature 387 733-736
[9]. Nadal M, et al. 2017 Nat. Commun. 8 14388
[10]. Thakur A, et al. 2024 Commun. Biol. 7 144
[11]. Hu L, et al. 2015 Nature 527 118-122
[12]. Ni Z, Lu W, Li Q, Han C, Yuan T, Sun N and Shi Y 2021 Cancer Biol. Med. 18 530-546
[13]. Zhang X, et al. 2019 Redox Biol. 24 101211
[14]. Jing T, Wei D, Xu X, Wu C, Yuan L, Huang Y, Liu Y, Jiang Y and Wang B 2024 Nat. Commun. 15 5631
[15]. Haque E, Teeli AS, Winiarczyk D, Taguchi M, Sakuraba S, Kono H, Leszczyński P, Pierzchała M and Taniguchi H 2022 Genes (Basel) 13 413
[16]. Aydin Y, et al. 2019 Cancers (Basel) 11 1407