1. Introduction
RNA-guided nucleases have been utilised by the CRISPR/Cas9 system, a heritable adaptive antiviral immune system of prokaryotes, to identify and destroy infectious invader viruses and bacteriophages. It includes two compartments: one for single-stranded guide RNA (sgRNA) and one for Cas9 endonuclease. The sgRNA directs the Cas9 endonuclease to specifically target the two DNA strands of the target gene for cleavage. DNA cleavage occurs 3 base pairs upstream of a "NGG" protospacer adjacent motif (PAM). Following the cleavage, the genome's DNA is restored via DNA-DSB repair procedures. Therefore, the extremely error-prone non-homologous end-joining (NHEJ) or the high-fidelity homology-directed repair (HDR) methods are utilised in the CRISPR/Cas9 gene editing system to add minuscule insertions or deletions (indels) to genomes (Figure 1).
One of the methods frequently employed in medical and pharmaceutical research is gene editing technology employing the Clustered Regularly Interspaced Short Palindromic Repeats and CRISPR-Associated Protein 9 System (CRISPR/Cas9). In order to test the impact of a gene's insertion or deletion on the levels of expression of the linked molecules, scientists can use genome editing to specifically knock out putative disease-causing genes. The three stages of the CRISPR/Cas9 technique are recognition, cleavage, and repair (Figure 1). Firstly, the targeted complementary sequence's protospacer adjacent motif (PAM) is recognized by the single guide RNA (sgRNA). The Cas9 nuclease then cleaves the target genome and causes a double strand break (DSB), under the control of sgRNA. Following the cleavage, there would be two different DSB repair mechanisms in action: homology-directed repair (HDR) and non-homologous end joining (NHEJ), which might result in precise editing or insertion/deletion, respectively[1].
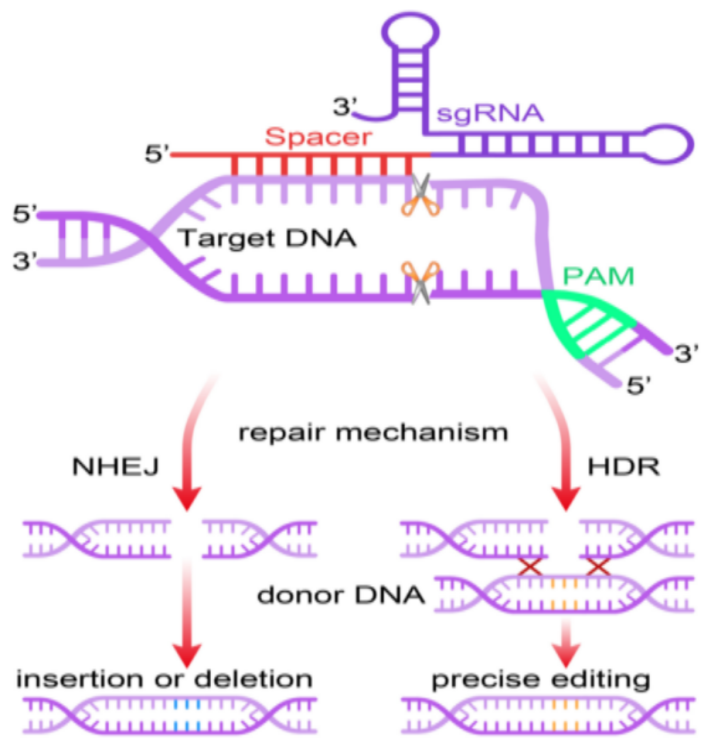
Figure 1. Overview of the CRISPR/Cas 9 genome editing technology.
Note. The CRISPR/Cas9 editing procedures are briefly illustrated in the figure above.[1]
In the past decade, the advancement of CRISPR technology has revolutionised genome engineering while making it a potent tool in the study of various diseases, from fundamental research to clinical trials and accurate treatment. The immune system destroys the myelin sheath in the central nervous system (CNS) in the autoimmune demyelinating illness that is known as multiple sclerosis (MS), exposing neurons and disrupting the transmission of electrical signals[2]. As a result, MS patients may experience a range of symptoms, including physical impairment and psychiatric issues, which can drastically lower their quality of life.
To find more effective treatments for MS, it is important to comprehend the disease states, such as the pathophysiological processes involved and the genetic alterations that lead to MS. Traditionally, oligodendrocyte precursor cell (OPC) culture, EAE rat models, and biochemical techniques that gauge cytokine levels, including the enzyme-linked immunosorbent assay (ELISA), have been utilized by scientists to investigate the disease states of MS[3-5]. However, these techniques have serious flaws, particularly the insufficient MS representation coverage.
The development of novel treatments for MS has benefited recently from the availability of new and creative tools to identify and validate drug targets. Many of these technologies are currently being used in MS research, such as DNA microarray analysis[6], which looks at alterations in MS-related miRNAs, and metabolomics[7] which tracks metabolic dysregulation in MS patients. CRISPR/Cas9 genomic editing technology has grown in prominence among these emerging technologies in the field of MS research.
2. Crispr/Cas9 applied on multiple sclerosis drug target validation advantages and disadvantages
The CRISPR/Cas9 technique has been used in a variety of research to investigate Multiple Sclerosis risk-associated DNA sequences. Based on isogenic cell lines with various genotypes altered by the CRISPR/Cas9 method, numerous prior investigations have discovered the impacts and locations of some MS-related causal SNPs and gene targets[8]. The effectiveness of increased protein expression on particular receptors has also been determined using this method employing KO mice models, such as the increased fibrinogen density's ability to mitigate OPC remyelination[9].
2.1. Advantages
The increased usage of CRISPR/Cas9 in MS medication research could be attributed to a number of benefits. Firstly, CRISPR/Cas9 permits high-throughput analysis of MS-causing gene activity, which may improve the effectiveness of translational research in preclinical investigations[10]. Secondly, by developing more humanised transgenic mouse models, this technique also addresses the issue of the absence of human analogous illness portrayal in animal models. Thirdly, by utilising fewer animals to produce genetic mouse models, CRISPR/Cas9 helps the 3Rs (Reduce, Reuse, and Recycle)[11]. Whenever MS-causing genes are evaluated functionally using conventional techniques like electrophoretic mobility shift tests, they are often isolated from their native genomic environment. By allowing access to the genes in their natural positions, CRISPR/Cas9 genome editing can get over this restriction and aid in the investigation of genes involved in the pathophysiology of MS[12]. Consequently, these benefits thus have the potential for significantly improving the efficacy of the creation of novel MS therapies.
2.2. Disadvantages
However, there are several restrictions that prevent wider adoption of CRISPR/Cas9. First of all, as CRISPR/Cas9 is typically employed to modify genes within the setting of a single kind of human or mouse cell, in vitro CRISPR/Cas9 models would not completely address the systemic immunological interactions involved in CNS inflammation in MS[10]. Furthermore, CRISPR/Cas9 may produce off-target effects. To ensure quick detection, researchers frequently build the sgRNA to recognize a brief motif called PAM on the targeted segments. The PAM segments may also appear in genomic regions that are unrelated to the target and unintended, leading to unexpected cleavage and alterations[13]. Additionally, Rodrigues-Rodriguez[14] link the risk of unintended mutations to the lack of knowledge of the DNA repair mechanisms that follow the use of CRISPR-Cas9. Moreover, this has rekindled the ethical debate regarding tampering with the human germ line. Despite the debate about "designer babies," CRISPR/Cas 9 provides up new possibilities for improving disease resistance in adults or fixing grave genomic errors in human embryos.Therefore, leading scientists have asked for a voluntary moratorium on human germ line genome editing until the ethical and scientific ramifications of doing so have been thoroughly examined. Two parties are locked in a "go/no-go" deadlock in the argument. One side argues that research into altering the human germ line should continue for the sake of science and medicine, while the other side argues that such editing is either immoral or harmful. However whether or not CRISPR/Cas 9 should be used to modify human germinal cells and embryos is not the most urgent ethical question that has to be answered right immediately. Already, human treatments are being created using CRISPR/Cas 9 to alter insects, animals, plants, and microbes. It might not appear that applying CRISPR/Cas 9 technology in these circumstances creates any new ethical concerns given that similar work has been done for years or even decades. However, there is the risk that CRISPR/Cas 9's efficiency and affordability will override long-standing, valid concerns about the production and spread of genetically modified organisms (GMOs). As demonstrated by the recent characterization of a brand-new type 2 CRISPR/Cas 9 system from Francisella novicida, the variety of genome editing techniques is continuously expanding.Therefore, there is an urgent need for comprehensive, worldwide regulations which regulate the testing of and release of GMOs into the environment[15]. Lastly, given that Cas9 is a nuclease derived from bacteria, human immune systems may create antibodies against it, which could limit the usefulness of the CRISPR/Cas9 system[13].
3. Discussion
A promising technique that allows for gene alteration to assist in locating or validating potential targets for MS is CRISPR/Cas9. Inv estigating the genes thought to be involved in MS aetiology and disease presentation is made possible by this method. This method could be utilised specifically to investigate whether MS-related pathways are up- or down-regulated in response to the knockout of particular genes[9]. Comparing the emerging method to conventional methods reveals several advantages. Because rodent OPCs and EAE animal models have biological differences, using CRISPR/Cas9 in human cell cultures has been able to overcome this restriction of insufficient accuracy[16]. Moreover, the CRISPR/Cas9 technique offers more precise and quantitative data that may be used for analysis in comparison to more typical phenotypic findings, such as the progression of impairment in EAE mouse models. However, compared to conventional procedures, there are a number of limitations to take into account. Since CRISPR/Cas9 entails irreversible genetic editing, it presents more serious ethical problems than conventional techniques[13]. Furthermore, CRISPR/Cas9 is less accessible since it requires advanced technical knowledge to preserve the specificity and precision of data, in contrast to the straightforward method in research employing the OPC and EAE models. Future MS research may apply these approaches in a variety of different directions. The hypothesised MS-linking genes could be completely and permanently knocked off using CRISPR/Cas9 editing in combination with additional methods.
For example, further positive-negative alternatives based on fluorescence display in Ikeda's team's[17] two-step genome editing strengthened the knockout stability, which was designed to allow for the editing of several genes. In my opinion, there are potential benefits to using CRISPR/Cas9 gene editing tools to validate therapeutic targets, such as MS targets. Studies utilising CRISPR/Cas9 gene editing tools may examine the makeup of specific experimental samples, such as sera and CSF. This opens the door to the potential of individualised therapy based on an examination of a person's protein and genetic profiles. The technique may also enhance the precision and effectiveness of MS target validation. The validation procedure for MS targets is accelerated by the use of the CRISPR/Cas9 gene editing method. Utilising cellular or animal models, CRISPR/Cas9 gene editing approaches could verify the functional relevance of the gene. On the contrary, a number of factors could stop the target validating progression. The extremely complicated gene structure associated with the technology known as CRISPR/Cas9 presents the greatest obstacles since it might be mislead and matched against different non-target sequences[18]. Fortunately, Cas9 protein modification or the use of alternative nucleases could increase editing precision[14].
4. Conclusion
The development of CRISPR technology over the past several decades has made precise and varied genome alteration possible. These adaptable technologies, that researchers currently refer to as "universal tools," revolutionised the life sciences and made basic research discoveries possible for an extensive variety of applications. The application of CRISPR in health care facilities is anticipated to provide a variety of therapeutic options for the treatment of human diseases, for instance multiple sclerosis. These technologies can be applied to several therapeutic applications if research into improving and revolutionising new methods of delivering genome engineering tools into cells and advancing their ability to edit continues. In MS research, CRISPR/Cas systems are widely used for a variety of in vitro and in vivo applications. A number of clinical trials are now being conducted to speed up or improve the medicines' dependability in order to effectively treat MS. However, these technologies should be created and placed into use in clinics, which needs continual, rigorous research. These technologies have the potential to advance in the present medical era and provide several alternatives for accurate and desired genome editing. By continuing to understand every shortcoming of the CRISPR system, improve editing capabilities, and create delivery methods, the complete ability of the CRISPR system to benefit society in the near future will be secured.
Acknowledgment
This article is written by Dongxu Qin, under the guide of professor Gordon Rule from Carnegie Mellon University and professor Dezhong Zhou from Xi’an Jiaotong University. Some suggestions from Xianrui Zhang from Peking University are also contributed to this article. During the writing time, Monash University helped finish the research and supply the research resources. Thanks to all the contributors.
References
[1]. Zhang, H., Qin, C., An, C., Zheng, X., Wen, S., Chen, W., Liu, X., Lv, Z., Yang, P., Xu, W., Gao, W., & Wu, Y. (2021). Application of the CRISPR/Cas9-based gene editing technique in basic research, diagnosis, and therapy of cancer. Molecular Cancer, 20, 126. https://doi.org/10.1186/s12943-021-01431-6
[2]. Dobson, R., & Giovannoni, G. (2018). Multiple sclerosis - a review. European Journal of Neurology, 26(1), 27–40. https://doi.org/10.1111/ene.13819
[3]. Wilson, H. C., Onischke, C., & Raine, C. S. (2003). Human oligodendrocyte precursor cells in vitro: Phenotypic analysis and differential response to growth factors. Glia, 44(2), 153–165. https://doi.org/10.1002/glia.10280
[4]. Wekerle, H. (2008). Lessons from multiple sclerosis: models, concepts, observations. Annals of the Rheumatic Diseases, 67(3), iii56–iii60. https://doi.org/10.1136/ard.2008.098020
[5]. Petzold, A., & Shaw, G. (2007). Comparison of two ELISA methods for measuring levels of the phosphorylated neurofilament heavy chain. Journal of Immunological Methods, 319(1-2), 34–40. https://doi.org/10.1016/j.jim.2006.09.021
[6]. Irizar, H., Muñoz-Culla, M., Sáenz-Cuesta, M., Osorio-Querejeta, I., Sepúlveda, L., Castillo-Triviño, T., Prada, A., Lopez de Munain, A., Olascoaga, J., & Otaegui, D. (2015). Identification of ncRNAs as potential therapeutic targets in multiple sclerosis through differential ncRNA – mRNA network analysis. BMC Genomics, 16(1), 250. https://doi.org/10.1186/s12864-015-1396-5
[7]. Zahoor, I., Rui, B., Khan, J., Datta, I., & Giri, S. (2021). An emerging potential of metabolomics in multiple sclerosis: a comprehensive overview. Cellular and Molecular Life Sciences, 78(7), 3181–3203. https://doi.org/10.1007/s00018-020-03733-2
[8]. Lee, M. H., Shin, J. I., Yang, J. W., Lee, K. H., Cha, D. H., Hong, J. B., Park, Y., Choi, E., Tizaoui, K., Koyanagi, A., Jacob, L., Park, S., Kim, J. H., & Smith, L. (2022). Genome Editing Using CRISPR-Cas9 and Autoimmune Diseases: A Comprehensive Review. International Journal of Molecular Sciences, 23(3), 1337. https://doi.org/10.3390/ijms23031337
[9]. Petersen, M. A., Ryu, J. K., Chang, K.-J., Etxeberria, A., Bardehle, S., Mendiola, A. S., Kamau-Devers, W., Fancy, S. P. J., Thor, A., Bushong, E. A., Baeza-Raja, B., Syme, C. A., Wu, M. D., Rios Coronado, P. E., Meyer-Franke, A., Yahn, S., Pous, L., Lee, J. K., Schachtrup, C., Lassmann, H., Huang, E. J., Han, M. H., Absinta, M., Reich, D. S., Ellisman, M. H., Rowitch, D. H., Chan, J. R., & Akassoglou, K. (2017). Fibrinogen Activates BMP Signaling in Oligodendrocyte Progenitor Cells and Inhibits Remyelination after Vascular Damage. Neuron, 96(5), 1003-1012.e7. https://doi.org/10.1016/j.neuron.2017.10.008
[10]. Johansen, K. H. (2021). How CRISPR/Cas9 Gene Editing Is Revolutionizing T Cell Research. DNA and Cell Biology, 41(1), 53–57. https://doi.org/10.1089/dna.2021.0579
[11]. Kirk, R. G. W. (2017). Recovering The Principles of Humane Experimental Technique. Science, Technology, & Human Values, 43(4), 622–648. https://doi.org/10.1177/0162243917726579
[12]. Ustiugova, A. S., Dvorianinova, E. M., Melnikova, N. V., Dmitriev, A. A., Kuprash, D. V., & Afanasyeva, M. A. (2023). CRISPR/Cas9 genome editing demonstrates functionality of the autoimmunity-associated SNP rs12946510. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease, 1869(4), 166599. https://doi.org/10.1016/j.bbadis.2022.166599
[13]. Yang, Y., Xu, J., Ge, S., & Lai, L. (2021). CRISPR/Cas: Advances, Limitations, and Applications for Precision Cancer Research. Frontiers in Medicine, 8, 649896. https://doi.org/10.3389/fmed.2021.649896
[14]. RodriguezRodriguez, D., RamirezSolis, R., GarzaElizondo, M., GarzaRodriguez, M., & BarreraSaldaia, H. (2019). Genome editing: A perspective on the application of CRISPR/Cas9 to study human diseases (Review). International Journal of Molecular Medicine, 43(4), 1559–1574. https://doi.org/10.3892/ijmm.2019.4112
[15]. Caplan, A. L., Parent, B., Shen, M., & Plunkett, C. (2015). No time to waste--the ethical challenges created by CRISPR. EMBO Reports, 16(11), 1421–1426. https://doi.org/10.15252/embr.201541337
[16]. Madill, M., Fitzgerald, D., O’Connell, K. E., Dev, K. K., Shen, S., & FitzGerald, U. (2016). In vitro and ex vivo models of multiple sclerosis. Drug Discovery Today, 21(9), 1504–1511. https://doi.org/10.1016/j.drudis.2016.05.018
[17]. Ikeda, K., Uchida, N., Nishimura, T., White, J., Martin, R. M., Nakauchi, H., Sebastiano, V., Weinberg, K. I., & Porteus, M. H. (2018). Efficient scarless genome editing in human pluripotent stem cells. Nature Methods, 15(12), 1045–1047. https://doi.org/10.1038/s41592-018-0212-y
[18]. Elliott, E. K., Haupt, L. M., & Griffiths, L. R. (2021). Mini review: genome and transcriptome editing using CRISPR-cas systems for haematological malignancy gene therapy. Transgenic Research, 30(2), 129–141. https://doi.org/10.1007/s11248-020-00232-9
Cite this article
Qin,D. (2024). Application of CRISPR/Cas9 on multiple sclerosis target validation. Theoretical and Natural Science,72,21-26.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 4th International Conference on Biological Engineering and Medical Science
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Zhang, H., Qin, C., An, C., Zheng, X., Wen, S., Chen, W., Liu, X., Lv, Z., Yang, P., Xu, W., Gao, W., & Wu, Y. (2021). Application of the CRISPR/Cas9-based gene editing technique in basic research, diagnosis, and therapy of cancer. Molecular Cancer, 20, 126. https://doi.org/10.1186/s12943-021-01431-6
[2]. Dobson, R., & Giovannoni, G. (2018). Multiple sclerosis - a review. European Journal of Neurology, 26(1), 27–40. https://doi.org/10.1111/ene.13819
[3]. Wilson, H. C., Onischke, C., & Raine, C. S. (2003). Human oligodendrocyte precursor cells in vitro: Phenotypic analysis and differential response to growth factors. Glia, 44(2), 153–165. https://doi.org/10.1002/glia.10280
[4]. Wekerle, H. (2008). Lessons from multiple sclerosis: models, concepts, observations. Annals of the Rheumatic Diseases, 67(3), iii56–iii60. https://doi.org/10.1136/ard.2008.098020
[5]. Petzold, A., & Shaw, G. (2007). Comparison of two ELISA methods for measuring levels of the phosphorylated neurofilament heavy chain. Journal of Immunological Methods, 319(1-2), 34–40. https://doi.org/10.1016/j.jim.2006.09.021
[6]. Irizar, H., Muñoz-Culla, M., Sáenz-Cuesta, M., Osorio-Querejeta, I., Sepúlveda, L., Castillo-Triviño, T., Prada, A., Lopez de Munain, A., Olascoaga, J., & Otaegui, D. (2015). Identification of ncRNAs as potential therapeutic targets in multiple sclerosis through differential ncRNA – mRNA network analysis. BMC Genomics, 16(1), 250. https://doi.org/10.1186/s12864-015-1396-5
[7]. Zahoor, I., Rui, B., Khan, J., Datta, I., & Giri, S. (2021). An emerging potential of metabolomics in multiple sclerosis: a comprehensive overview. Cellular and Molecular Life Sciences, 78(7), 3181–3203. https://doi.org/10.1007/s00018-020-03733-2
[8]. Lee, M. H., Shin, J. I., Yang, J. W., Lee, K. H., Cha, D. H., Hong, J. B., Park, Y., Choi, E., Tizaoui, K., Koyanagi, A., Jacob, L., Park, S., Kim, J. H., & Smith, L. (2022). Genome Editing Using CRISPR-Cas9 and Autoimmune Diseases: A Comprehensive Review. International Journal of Molecular Sciences, 23(3), 1337. https://doi.org/10.3390/ijms23031337
[9]. Petersen, M. A., Ryu, J. K., Chang, K.-J., Etxeberria, A., Bardehle, S., Mendiola, A. S., Kamau-Devers, W., Fancy, S. P. J., Thor, A., Bushong, E. A., Baeza-Raja, B., Syme, C. A., Wu, M. D., Rios Coronado, P. E., Meyer-Franke, A., Yahn, S., Pous, L., Lee, J. K., Schachtrup, C., Lassmann, H., Huang, E. J., Han, M. H., Absinta, M., Reich, D. S., Ellisman, M. H., Rowitch, D. H., Chan, J. R., & Akassoglou, K. (2017). Fibrinogen Activates BMP Signaling in Oligodendrocyte Progenitor Cells and Inhibits Remyelination after Vascular Damage. Neuron, 96(5), 1003-1012.e7. https://doi.org/10.1016/j.neuron.2017.10.008
[10]. Johansen, K. H. (2021). How CRISPR/Cas9 Gene Editing Is Revolutionizing T Cell Research. DNA and Cell Biology, 41(1), 53–57. https://doi.org/10.1089/dna.2021.0579
[11]. Kirk, R. G. W. (2017). Recovering The Principles of Humane Experimental Technique. Science, Technology, & Human Values, 43(4), 622–648. https://doi.org/10.1177/0162243917726579
[12]. Ustiugova, A. S., Dvorianinova, E. M., Melnikova, N. V., Dmitriev, A. A., Kuprash, D. V., & Afanasyeva, M. A. (2023). CRISPR/Cas9 genome editing demonstrates functionality of the autoimmunity-associated SNP rs12946510. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease, 1869(4), 166599. https://doi.org/10.1016/j.bbadis.2022.166599
[13]. Yang, Y., Xu, J., Ge, S., & Lai, L. (2021). CRISPR/Cas: Advances, Limitations, and Applications for Precision Cancer Research. Frontiers in Medicine, 8, 649896. https://doi.org/10.3389/fmed.2021.649896
[14]. RodriguezRodriguez, D., RamirezSolis, R., GarzaElizondo, M., GarzaRodriguez, M., & BarreraSaldaia, H. (2019). Genome editing: A perspective on the application of CRISPR/Cas9 to study human diseases (Review). International Journal of Molecular Medicine, 43(4), 1559–1574. https://doi.org/10.3892/ijmm.2019.4112
[15]. Caplan, A. L., Parent, B., Shen, M., & Plunkett, C. (2015). No time to waste--the ethical challenges created by CRISPR. EMBO Reports, 16(11), 1421–1426. https://doi.org/10.15252/embr.201541337
[16]. Madill, M., Fitzgerald, D., O’Connell, K. E., Dev, K. K., Shen, S., & FitzGerald, U. (2016). In vitro and ex vivo models of multiple sclerosis. Drug Discovery Today, 21(9), 1504–1511. https://doi.org/10.1016/j.drudis.2016.05.018
[17]. Ikeda, K., Uchida, N., Nishimura, T., White, J., Martin, R. M., Nakauchi, H., Sebastiano, V., Weinberg, K. I., & Porteus, M. H. (2018). Efficient scarless genome editing in human pluripotent stem cells. Nature Methods, 15(12), 1045–1047. https://doi.org/10.1038/s41592-018-0212-y
[18]. Elliott, E. K., Haupt, L. M., & Griffiths, L. R. (2021). Mini review: genome and transcriptome editing using CRISPR-cas systems for haematological malignancy gene therapy. Transgenic Research, 30(2), 129–141. https://doi.org/10.1007/s11248-020-00232-9









