1.Introduction
1.1.Calcium Imaging
Calcium Imaging, a powerful technique in the field of neuroscience that allows researchers to observe and study the activity of individual neurons or populations of neurons by monitoring changes in intracellular calcium ion concentrations [1,2]. Calcium ions play a crucial role in various cellular processes, including neurotransmitter release, synaptic plasticity, and intracellular signal [3]. The technique involves the use of fluorescent calcium indicators, which are molecules that emit fluorescent light when they bind to calcium ions. These indicators can be introduced into neurons either through genetic manipulation (such as expressing a genetically encoded calcium indicator) or by loading them into the cells using specialized dyes [4,5]. When a neuron becomes active and experiences an increase in intracellular calcium levels—often due to action potentials or synaptic input—the calcium indicators bind to the calcium ions and emit fluorescence in response [3]. By capturing this fluorescent signal, researchers can visualize and record neuronal activity in real-time. In this investigation, the fluorescent signal data originates from GCaMP, one of the indicators researchers applying for.
1.2.GCaMP
GCaMP (Genetically Encoded Calcium Indicator) fluorescence is a widely used tool in neuroscience for monitoring changes in intracellular calcium levels in living cells, particularly neurons [6]. GCaMP is a genetically engineered protein that undergoes a change in fluorescence intensity when it binds to calcium ions [6]. This change in fluorescence provides a real-time readout of neuronal activity, making it a valuable method for studying neural processes and understanding the dynamics of neuronal networks [7].
In this research, it analyzes and processes the GCaMP fluorescence recording data to infer the possible underlying spikes by using Online Active Set method to Infer Spike (OASIS), a state-of-the-art deconvolution technique inspired by the pool adjacent violators algorithm (PAVA) [8], It establishes a method to realize a fast real-time data processing, because while the rise and decay time of GCaMP is in the order of hundreds of milliseconds, it is impossible to observe the exact single action potentials which occurs in just a few milliseconds [8]. The GCaMP6 fluorescence recording data sets is mouse primary visual cortex including time series traces of hundreds of mouse visual cortex(V1) neurons. However, before inferring the underlying spikes, GCaMP fluorescence recording data should be filtered due to its low-frequency noise, which contains animal motion, photobleaching and imaging noise, compared with relatively high-frequency of GCaMP fluorescence. Basically, the data be processed by applying high-pass filter to eliminate the noise baseline drift.
1.3.OASIS
The Online Active Set method to Infer Spike (OASIS) is a state-of-the-art deconvolution technique employed in neuroscience for inferring neuronal spikes from fluorescence recording data [8]. OASIS operates based on a principle analogous to the pool adjacent violators algorithm (PAVA), which optimally estimates the underlying spike train given the observed fluorescence signal [8]. OASIS functions by iteratively updating a sparse binary matrix that denotes spike occurrences over time [8]. The algorithm seeks to find a set of spike times that best reconstructs the observed fluorescence signal while adhering to the inherent temporal dynamics of neuronal activity [8]. This involves solving a convex optimization problem that minimizes the difference between the observed fluorescence trace and a reconstructed spike train convolved with a modeled calcium response function [8]. At each iteration, OASIS identifies a potential active set of spikes and computes their contributions to the fluorescence signal [8]. The algorithm evaluates the goodness-of-fit of this active set and adjusts it to better match the observed data [8]. This iterative refinement process continues until a satisfactory match between the inferred spike train and the observed fluorescence is achieved. OASIS demonstrates its strength in its ability to operate in real-time, making it particularly valuable for studying neural processes with high temporal resolution [8]. Its capacity to reliably infer spike times from noisy fluorescence data contributes to the understanding of neuronal dynamics, facilitating insights into information processing, neural coding, and network interactions [8].
2.Methodology
To filter the GCaMP fluorescence signal data, firstly, MATLAB was opened and the GCaMP fluorescence data which were\( N×M \)matrix of GCaMP\( \frac{∆F}{{F_{0}}} \)(\( {F_{0}} \)was baseline fluorescence value,\( ∆F \)was relative change) were averaged by column. Then, the data were Fourier transformed and the average Fourier magnitude spectrum was plotted. Next, the cutoff frequency of a suitable ideal high-pass filter was determined based on the average Fourier magnitude spectrum. Then, a discrete high-pass filter was designed by the cutoff frequency, the length of data and the frame rate. Subsequently, the inverse Fourier transform was applied to the discrete high-pass filter to be the high-pass filter coefficients. Then the original mean data and the high-pass filter transformed by the inverse Fourier transform were cyclically convolved. The result was plotted as the filtered data, and its Fourier-transformed image was also plotted. After obtaining the processed standard GCaMP fluorescence time series data, the corrcoef equation was applied to the filtered data sets to find their connectivity.
To infer the GCaMP fluorescence signal data spike fast and in real-time, first of all, the OASIS GitHub page was opened and the OASIS package was downloaded. Then, the main function in the OASIS package was directly referenced, the original data were deconvolved through the main function in the OASIS package, and the resulting image was drawn. Finally, the value greater than 0 was selected as a new array, and the firing rate was obtained by dividing the length of the new array by the original data and multiplying by the frame rate.
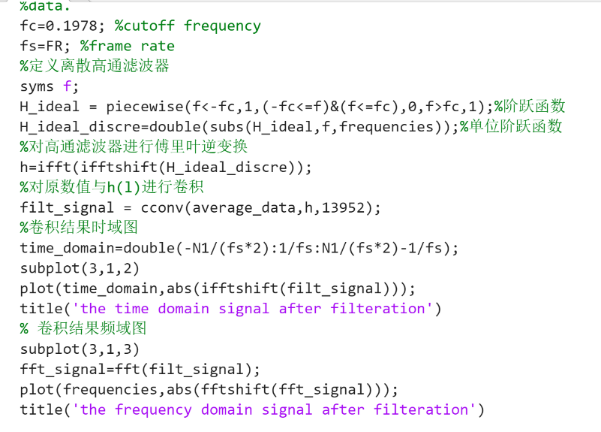
Figure 1. The Code of Plotting Data after being Filtered.
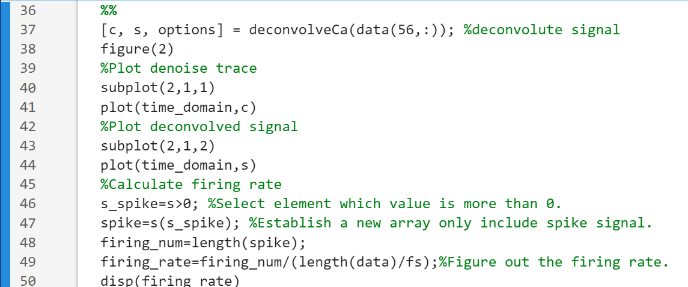
Figure 2. The Code of Inferring Spikes for Raw Data.
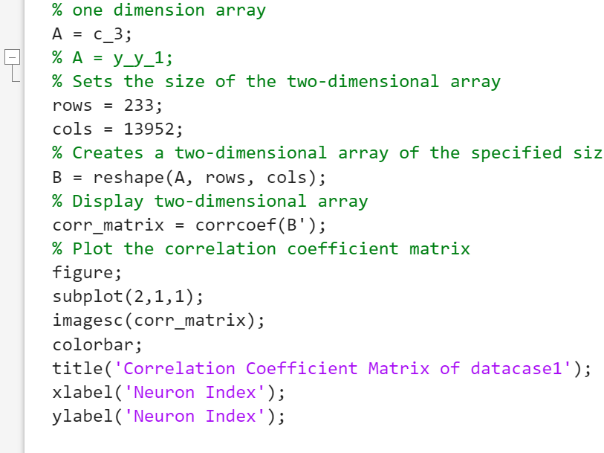
Figure 3. The Code for Correlation Coefficient of Data.
3.Result and discussion
The results and discussion section presents the outcomes of the research:
Cutoff Frequency Selection and filtered signal: The cutoff frequency of 0.1978 is chosen for the high-pass filter, effectively filtering out low-frequency noise while retaining high-frequency signals.
Inference of Neuronal Spikes: The OASIS package successfully infers neuronal spikes from the processed GCaMP fluorescence data. Figure 4 visually represents the inferred spikes, and the calculated firing rate is 0.4125.
Further Possibilities: The paper suggests that other filtering techniques like bandpass filters could be explored for noise reduction. Additionally, comparing data processed through the OASIS algorithm with Gaussian mixture models could provide more insights. Comparisons of data connectivity and centrality could contribute to further research.
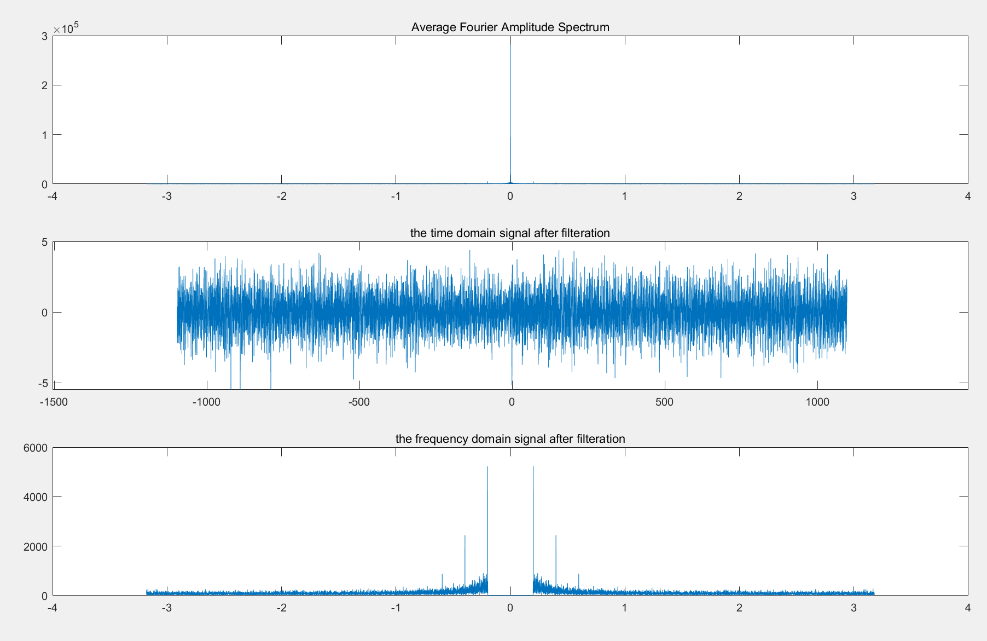
Figure 4. GCaMP Fluorescence Data after High-pass Filter Processing
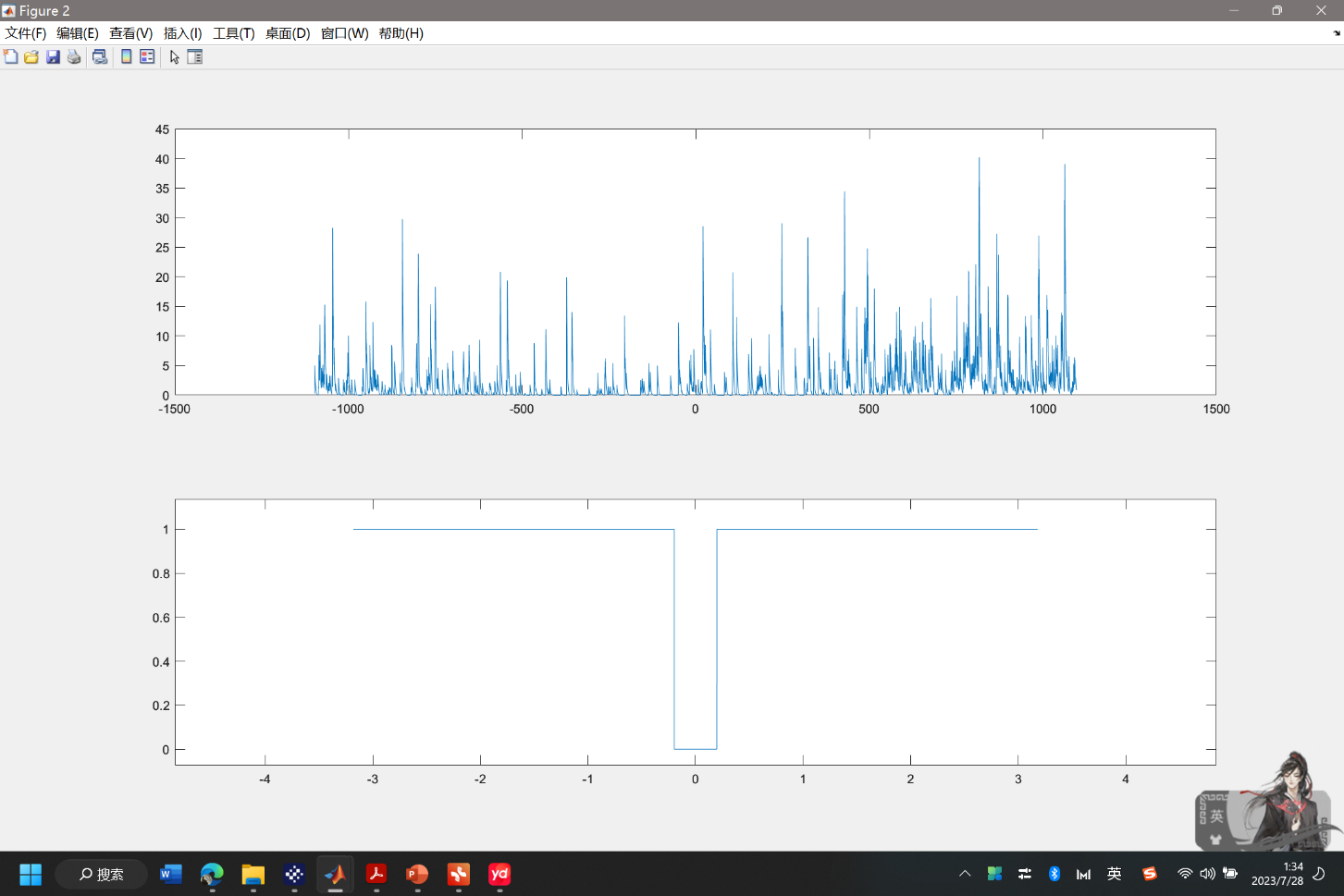
Figure 5. The Inferred Spike from GCaMP Fluorescence Data after Applying OASIS Algorithm.
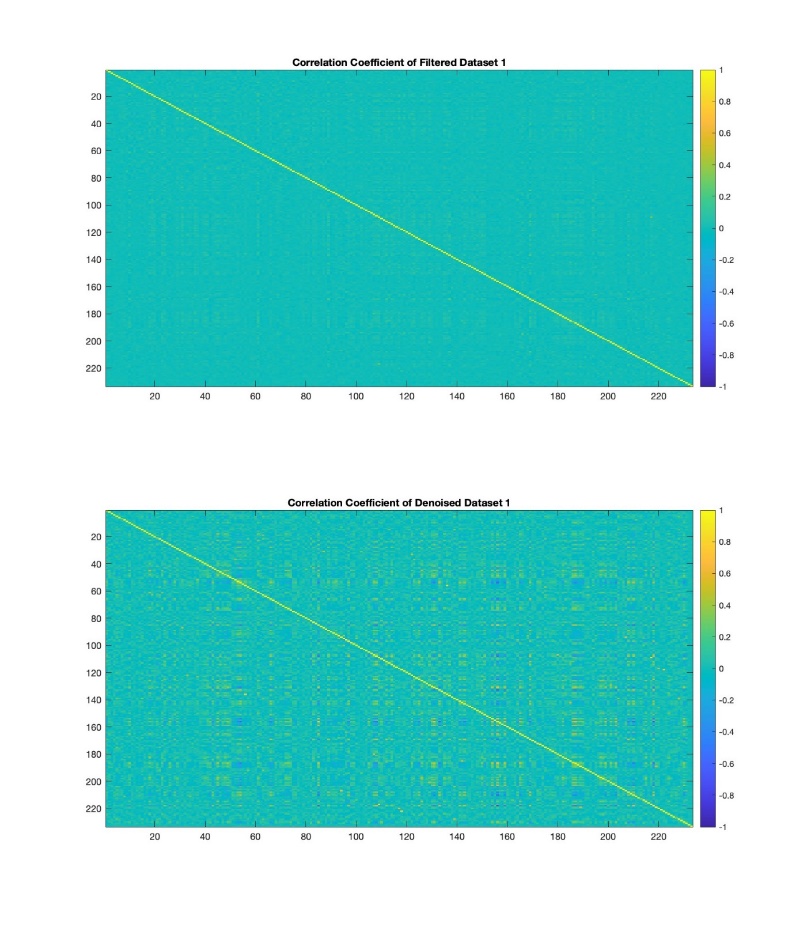
Figure 6. The Correlation Coefficient of Filtered and Denoised Data.
4.Conclusion
In summary, this study employed GCaMP fluorescence data which comprehensively examine neural activity in the mouse primary visual cortex. Utilizing GCaMP, which emits fluorescence upon binding with calcium ions, allowed real-time tracking of neural dynamics [3]. The purpose of research is GCaMP data preprocessing and the Online Active Set method to Infer Spike (OASIS).
High-pass filtering effectively removed low-frequency noise, and OASIS revealed neuronal spikes, inferring a frequency of 0.4125. Exploring alternative noise reduction methods like bandpass filtering and comparing results with Gaussian mixture models could.
This study presents a novel approach for real-time monitoring and inference of neural activity, extending possibilities for future neuroscientific research to further effectively remove noise and analyze OASIS algorithm.
References
[1]. Carsen Stringer, Marius Pachitariu, Nicholas Steinmetz, Charu Bai Reddy, Matteo Carandini, Kenneth D. Harris bioRxiv 306019. https://doi.org/10.1101/306019
[2]. Ahrens, M., Orger, M., Robson, D. et al. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat Methods 10, 413–420 (2013). https://doi.org/10.1038/nmeth.2434
[3]. Grienberger, Christine et al. Imaging Calcium in Neurons. Neuron, Volume 73, Issue 5, 862 – 885. https://doi.org/10.1016/j.neuron.2012.02.011
[4]. Palmer, A., Tsien, R. Measuring calcium signaling using genetically targetable fluorescent indicators. Nat Protoc 1, 1057–1065 (2006). https://doi.org/10.1038/nprot.2006.172
[5]. R. Madelaine Paredes, Julie C. Etzler, Lora Talley Watts, Wei Zheng, James D. Lechleiter. Chemical calcium indicators. Methods. Volume 46, Issue 3, 2008, Pages 143-151, ISSN 1046-2023. https://doi.org/10.1016/j.ymeth.2008.09.025
[6]. Crystal Structures of the GCaMP Calcium Sensor Reveal the Mechanism of Fluorescence Signal Change and Aid Rational Design *Akerboom, Jasper et al. Journal of Biological Chemistry, Volume 284, Issue 10, 6455 - 6464
[7]. Optimization of a GCaMP Calcium Indicator for Neural Activity Imaging. Jasper Akerboom, Tsai-Wen Chen, Trevor J. Wardill, Lin Tian, Jonathan S. Marvin, Sevinç Mutlu, Nicole Carreras Calderón, Federico Esposti, Bart G. Borghuis, Xiaonan Richard Sun, Andrew Gordus, Michael B. Orger, Ruben Portugues, Florian Engert, John J. Macklin, Alessandro Filosa, Aman Aggarwal, Rex A. Kerr, Ryousuke Takagi, Sebastian Kracun, Eiji Shigetomi, Baljit S. Khakh, Herwig Baier, Leon Lagnado, Samuel S.-H. Wang, Cornelia I. Bargmann, Bruce E. Kimmel, Vivek Jayaraman, Karel Svoboda, Douglas S. Kim, Eric R. Schreiter, Loren L. Looger. Journal of Neuroscience 3 October 2012, 32 (40) 13819-13840; doi: 10.1523/JNEUROSCI.2601-12.2012
[8]. Fast online deconvolution of calcium imaging data. Friedrich J, Zhou P, Paninski L (2017) Fast online deconvolution of calcium imaging data. PLOS Computational Biology 13(3): e1005423. https://doi.org/10.1371/journal.pcbi.1005423
Cite this article
Zhang,J. (2025). Signal Processing and Data Analysis for GCaMP Filtering and OASIS Algorithm. Theoretical and Natural Science,73,172-176.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 4th International Conference on Biological Engineering and Medical Science
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Carsen Stringer, Marius Pachitariu, Nicholas Steinmetz, Charu Bai Reddy, Matteo Carandini, Kenneth D. Harris bioRxiv 306019. https://doi.org/10.1101/306019
[2]. Ahrens, M., Orger, M., Robson, D. et al. Whole-brain functional imaging at cellular resolution using light-sheet microscopy. Nat Methods 10, 413–420 (2013). https://doi.org/10.1038/nmeth.2434
[3]. Grienberger, Christine et al. Imaging Calcium in Neurons. Neuron, Volume 73, Issue 5, 862 – 885. https://doi.org/10.1016/j.neuron.2012.02.011
[4]. Palmer, A., Tsien, R. Measuring calcium signaling using genetically targetable fluorescent indicators. Nat Protoc 1, 1057–1065 (2006). https://doi.org/10.1038/nprot.2006.172
[5]. R. Madelaine Paredes, Julie C. Etzler, Lora Talley Watts, Wei Zheng, James D. Lechleiter. Chemical calcium indicators. Methods. Volume 46, Issue 3, 2008, Pages 143-151, ISSN 1046-2023. https://doi.org/10.1016/j.ymeth.2008.09.025
[6]. Crystal Structures of the GCaMP Calcium Sensor Reveal the Mechanism of Fluorescence Signal Change and Aid Rational Design *Akerboom, Jasper et al. Journal of Biological Chemistry, Volume 284, Issue 10, 6455 - 6464
[7]. Optimization of a GCaMP Calcium Indicator for Neural Activity Imaging. Jasper Akerboom, Tsai-Wen Chen, Trevor J. Wardill, Lin Tian, Jonathan S. Marvin, Sevinç Mutlu, Nicole Carreras Calderón, Federico Esposti, Bart G. Borghuis, Xiaonan Richard Sun, Andrew Gordus, Michael B. Orger, Ruben Portugues, Florian Engert, John J. Macklin, Alessandro Filosa, Aman Aggarwal, Rex A. Kerr, Ryousuke Takagi, Sebastian Kracun, Eiji Shigetomi, Baljit S. Khakh, Herwig Baier, Leon Lagnado, Samuel S.-H. Wang, Cornelia I. Bargmann, Bruce E. Kimmel, Vivek Jayaraman, Karel Svoboda, Douglas S. Kim, Eric R. Schreiter, Loren L. Looger. Journal of Neuroscience 3 October 2012, 32 (40) 13819-13840; doi: 10.1523/JNEUROSCI.2601-12.2012
[8]. Fast online deconvolution of calcium imaging data. Friedrich J, Zhou P, Paninski L (2017) Fast online deconvolution of calcium imaging data. PLOS Computational Biology 13(3): e1005423. https://doi.org/10.1371/journal.pcbi.1005423









