1. Introduction
As a state-of-the-art modern treatment in life sciences study, stem cell therapy has emerged as world's most common specific therapy to the genetic diseases. Physicians can thus efficiently and safely treat the majority of patients with genetic diseases by targeting pathogenic mutations with somatic cell gene editing. In 2006, Shinya Yamanaka and his team achieved a breakthrough by importingOct4, Sox2, Klf4, and c-Myc into fibroblasts using viral vectors. Furthermore, this landmark discovery allowed for cells to take on such a pluripotent differentiation potential, which could then give rise to an iPSC. Back in 2007, Dr Kathrin Plath and her group were among the first to reprogramme skin cells into precursors of gametes, hepatocytes and skeletal cells using iPSCs. 18 19 iPSCs have unlimited proliferative potential and the capacity to differentiate into a range of homogeneous somatic cells, as well as tissues, making them highly applicable for cell therapy and tissue regeneration. On top of this, iPSCs bypass the ethical and immunogenic issues of embryonic stem cells [1].
CRISPR-Cas9 is regarded as a revolutionary gene-editing technology, which can assist scientists to modify genetic sequences more efficiently and precisely. CRISPR, a naturally occurring immune system found in bacteria and archaea, works in tandem with Cas9, a nuclease associated with the CRISPR system. By utilizing small RNA sequences complementary to target DNA, the system employs a guide RNA (sgRNA)—comprising crRNA and tracrRNA—to direct the Cas9 endonuclease to the desired DNA sequence. The Cas9 protein then induces double-strand breaks (DSBs) at specific target sites in the DNA. Subsequent DNA repair mechanisms can result in gene knockout or insertion. Compared to traditional homologous recombination methods, CRISPR-Cas9 offers significant advantages in speed and efficiency.
Applications of CRISPR-Cas9 include targeted gene editing, ranging from its initial role as a research tool to potential therapeutic interventions. CRISPR-Cas9 is gradually transitioning from a laboratory tool to clinical practice. For instance, Liu, et al. initiated clinical trials targeting gastric cancer and nasopharyngeal carcinoma, treating two gastric cancer patients with CRISPR-Cas9 [2]. On June 17, 2021, the UK reported the first clinical data demonstrating the safety and efficacy of in vivo CRISPR gene editing, showing that a single intravenous injection of CRISPR could precisely edit target cells within the body to treat genetic diseases.
Despite the high accuracy of CRISPR-Cas9 in targeting specific DNA sequences, off-target effects remain a concern, as unintended cuts in non-specific fragments could lead to cellular dysfunction or mutations, increasing safety risks. Consequently, CRISPR-Cas9 technology is evolving toward greater specificity. By integrating gene repair mechanisms—such as homologous recombination—following Cas9-induced DNA cleavage, the precision of DNA repair can be further enhanced, minimizing unintended effects and improving the overall safety profile.
2. The origins of CRISPR/Cas9
The progress of CRISPR/Cas9 is shown in Figure 1. When regularly spaced palindromic repeats were discovered in DNA by Japanese scientists of E. coli K-12. This structure, later recognized as a series of regular palindromic repeats interspaced with unique sequences, serves as a defense mechanism for archaea and other microorganisms against viruses and foreign genetic elements. In 2002, this system was formally named CRISPR, alongside its associated proteins (Cas), particularly Cas9.
Such a groundbreaking milestone was achieved in 2012, when the Cas9 nuclease was first identified to possess both RNA-guided and DNA-cleaving functions. Subsequently, in 2013, researchers successfully applied this system to the genetic editing of eukaryotic cells, marking a transformative leap in the field of life sciences.
As of 2023, regulatory authorities in the United Kingdom have approved a CRISPR-based gene-editing therapy known as Casgevy. This approval underscores the immense potential of CRISPR/Cas9 and highlights its promising applications across diverse fields, including therapeutic development and beyond [3].
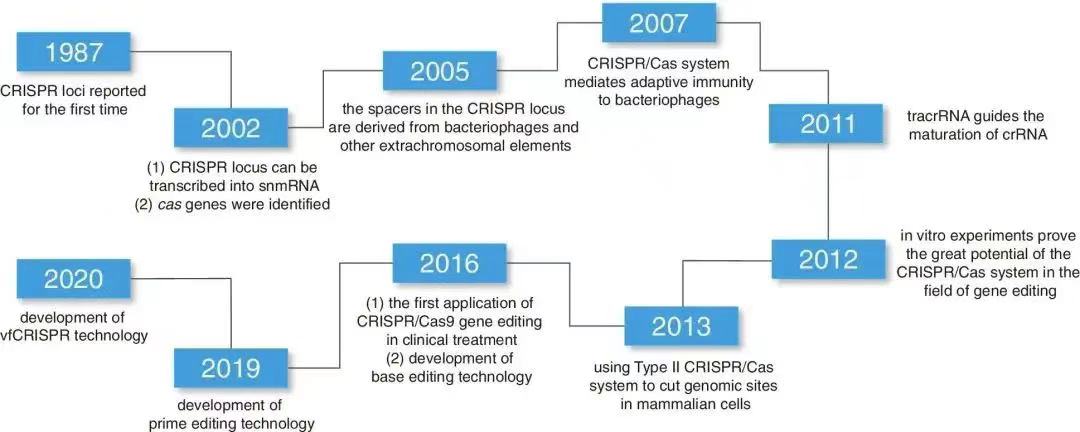
Figure 1: History of Development and the Evolution of Related Gene Editing Tools [4].
Gene editing relies on two key components. The Cas9 enzyme contains two functional domains, HNH and RuvC. The HNH domain cleaves the DNA strand targeted by the sgRNA, while the RuvC domain cleaves the non-target strand. Guided by sgRNA, Cas9 induces a DSB at the target gene. The break is then repaired through the cell's endogenous DNA repair pathways—either non-homologous end joining (NHEJ) or homologous recombination (HDR)—resulting in gene knockout, insertion, or mutation.
Studies on the Cas9 molecular structure reveal that Cas9 first binds to sgRNA, and under the guidance of sgRNA, it further associates with the target double-stranded DNA. A conformational change in Cas9 unwinds the target DNA, allowing the sgRNA strand to invade and specifically bind to the DNA strand complementary to the sgRNA. Mechanistic studies also show that the protospacer adjacent motif (PAM) sequence at the 3’ end of the target sequence is crucial for initial DNA binding. Without PAM, the target sequence, even if perfectly complementary to the sgRNA, will not be recognized by Cas9. Once sequence complementarity is achieved, Cas9 activates its nuclease activity. The HNH and RuvC domains each cleave one DNA strand, leading to the formation of a DNA DSB. By mutating either of these two domains, the Cas9 protein can be converted into a nickase (nCas9), which only cuts one strand. Mutating both domains results in a dead Cas9 (dCas9), which has no endonuclease activity. These two Cas9 variants, nCas9 and dCas9, are widely used for constructing various genetic manipulation tools [5].
Cas9-induced double-strand breaks (DSBs) are typically repaired by the cell's DNA repair pathways: non-homologous end joining (NHEJ) and homology-directed repair (HDR). NHEJ is an error-prone repair mechanism that rapidly joins the broken ends of DNA, but it often results in insertions or deletions (indels) at the break site, leading to frameshift mutations and knockout of the target gene. In contrast, HDR allows precise repair by introducing a donor template. By providing a repair template, HDR can accurately modify endogenous genes near the Cas9 cleavage site.
Application of CRISPR/Cas9 in Establishing Animal Models
Currently, CRISPR/Cas9 is primarily used to establish cellular and animal models. In the field of cancer research, various tumor cell lines and animal models based on the CRISPR/Cas9 system have been successfully developed and applied for preclinical validation in areas such as cancer genes, drug targets, and drug resistance [6].
The establishment of cancer animal models is a crucial tool for studying the functions of cancer-related genes, and CRISPR/Cas9 has significantly simplified the process of creating these models, reducing costs and accelerating modeling timelines. By using CRISPR/Cas9, researchers can study the roles of specific genes in tumor initiation and progression. Currently, CRISPR/Cas9-based tumor models have been established for various cancers. For example, one study used CRISPR/Cas9 to silence the expression of the CDK11 gene in osteosarcoma cell lines KHOS and U-2OS, and found that silencing CDK11 significantly reduced osteosarcoma cell proliferation and invasion. This discovery not only revealed the critical role of CDK11 in osteosarcoma, but also provided a theoretical foundation for the development of new therapy strategies.
The development of chronic myelogenous leukemia (CML) is often associated with mutations in the tumor suppressor gene ASXL1, which impacts patient prognosis. In another study, researchers used CRISPR/Cas9 to target and repair the ASXL1 mutation in KBM5 cells, significantly reducing the proliferation rate of the cells and enhancing their differentiation. Mice with ASXL1-corrected tumors had a longer survival time compared to those with untreated tumors. This study not only confirmed the pivotal role of ASXL1 in CML but also offered new perspectives for future gene therapy.
Additionally, researchers from the University of Cambridge and other institutions conducted large-scale CRISPR/Cas9 screening of 324 tumor cell lines derived from 30 different malignancies, developing a database called Project Score. This database helps identify potential therapeutic targets for cancer. It contains extensive functional annotations of genes, as well as detailed experimental data and analysis tools, enabling researchers to more efficiently identify potential therapeutic targets.
3. Chimeric Antigen Receptor (CAR)-T Cell Therapy
Since the FDA approved two autologous CAR-T in 2017, numerous clinical trials have demonstrated the efficacy of CAR-T in treating various hematologic and non-hematologic malignancies. However, clinical applications are still limited by certain challenges, and CRISPR genome editing offers a potential solution to address these limitations (Figure 2). Recent studies have highlighted that CRISPR-based therapies can be used to target CAR-T directed against CD19 and CD70, providing a promising approach for the treatment of solid tumors, leukemia, and lymphoma [7].
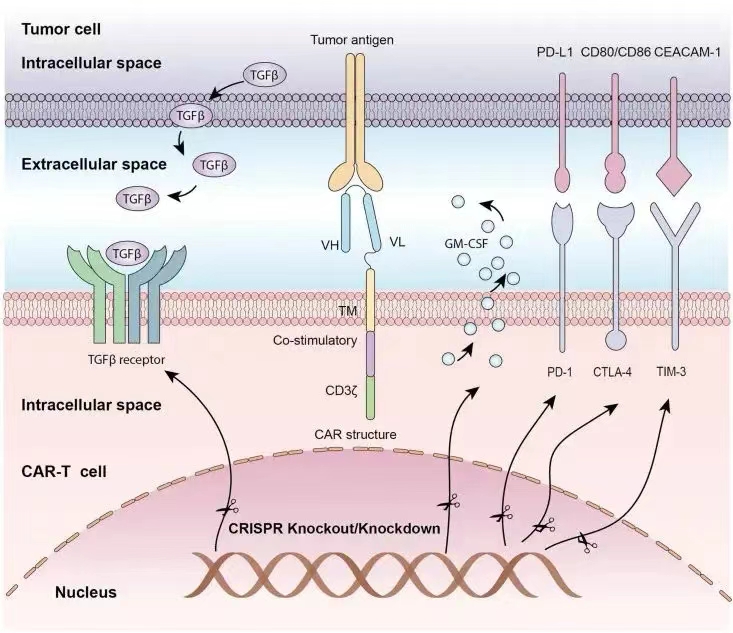
Figure 2: Application of the CRISPR/Cas9 System in CAR-T Cell Editing [8].
4. Transgenic T Cell Receptor (TCR)-T Cell Therapy
CRISPR/Cas9-mediated knockout of endogenous TCR-β in T cells, combined with the transduction of cancer-reactive receptors, significantly increases the expression of transgenic T cell receptors (tgTCR), enhancing the engineered T cells' ability to target and kill cancer cells. Compared to standard TCR-transduced T cells, TCR-transduced and CRISPR-edited T cells exhibit a thousand-fold greater sensitivity to the antigen [9].
The first clinical trial report of 2020 revealed the use of multiple CRISPR/Cas9 edits to target T cell genes in order to enhance anti-tumor immune responses. Additionally, synthetic cancer-specific TCR transgenes (which recognize tumor cells) were introduced. Engineered T cells persist in patients for up to 9 months, providing preliminary evidence that combining TCR transfer with genome editing may lead to more effective and safer cancer immunotherapies [10].
5. Conclusion
Significant progress has been made in stem cell therapy within current life sciences research, particularly in the treatment of major diseases such as genetic disorders and cancer. Through somatic gene editing technologies, scientists are now able to precisely repair or replace pathogenic genes, offering new therapeutic hope for patients. Moreover, breakthroughs in iPSC technology have made it possible to reprogram adult cells into pluripotent stem cells, providing abundant cellular sources for regenerative medicine and opening new avenues for disease model establishment and drug screening.
Despite the many achievements in stem cell therapy, several challenges remain. First, the safety and efficacy of gene editing still require further validation, especially regarding long-term effects and potential side effects. Additionally, the mechanisms governing stem cell differentiation are not yet fully understood, and efficiently and stably directing stem cells to differentiate into specific types of somatic cells remains a major focus of research. Ethical and legal issues also pose significant barriers to the widespread application of stem cell therapy.
To overcome these challenges, it is essential to optimize gene editing tools such as CRISPR/Cas9, reduce off-target effects, improve editing efficiency, and ensure the safety and reliability of treatments. Further research into the key signaling pathways and molecular mechanisms involved in stem cell differentiation is critical to developing more effective differentiation induction strategies, enabling precise and personalized cell therapies. Establishing a robust ethical review system and legal framework is crucial to ensure the regulatory and ethical integrity of stem cell research and applications, protecting patient rights and promoting the healthy development of the technology. We can accelerate the translation of basic research findings into clinical applications and conducting more human clinical trials are essential to validate the security and efficacy of stem cell therapies and deliver tangible benefits to patients.
In the future, iPSC is expected to make breakthroughs in several key areas: achieving precision medicine through genetic sequencing and individualized stem cell culture; combining gene editing, immunotherapy, and other technologies to enhance therapeutic efficacy; utilizing biomanufacturing technologies to enable large-scale production and standardized preparation of stem cells; and establishing comprehensive ethical review and regulatory mechanisms to safeguard patient rights.
References
[1]. Takahashi, K., & Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell, 126, 663-676.
[2]. Xu, R. L., et al. (2019). Correction of a pathogenic gene mutation in human embryos using CRISPR-Cas9.
[3]. Hochedlinger, K., & Plath, K. (2009). Epigenetic reprogramming and induced pluripotency. Development, 136, 509-523.
[4]. Wang, S. W., Gao, C., Zheng, Y. M., et al. (2022). Current applications and future perspective of CRISPR/Cas9 gene editing in cancer. Molecular Cancer, 21, 57.
[5]. Xiong, Z., & Jiang, Y. (2024). Research progress on methods to improve the efficacy of mesenchymal stem cell transplantation for treating liver cirrhosis. Journal of Gastroenterology and Hepatology, 33(10), 1401-1404.
[6]. Normile, D. (2017). China sprints ahead in CRISPR therapy race. Science, 358, 20-21.
[7]. Rupp, L. J., Schumann, K., Roybal, K. T., et al. (2017). CRISPR/Cas9-mediated PD-1 disruption enhances anti-tumor efficacy of human chimeric antigen receptor T cells. Scientific Reports, 7(1), 1-10.
[8]. Ou, X., Ma, Q., Yin, W., et al. (2021). CRISPR/Cas9 gene-editing in cancer immunotherapy: Promoting the present revolution in cancer therapy and exploring more. Frontiers in Cell and Developmental Biology, 9, 674467.
[9]. Feng, Y., Sassi, S., Shen, J. K., et al. (2015). Targeting Cdk11 in osteosarcoma cells using the CRISPR-Cas9 system. Journal of Orthopaedic Research, 33(2), 199-207.
[10]. Rath, J. A., & Arber, C. (2020). Engineering strategies to enhance TCR-based adoptive T cell therapy. Cells, 9(6), 1485.
Cite this article
Wu,H. (2025). Advances in the Application of CRISPR-Cas9 in Stem Cell Therapy. Theoretical and Natural Science,90,75-79.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of ICMMGH 2025 Workshop: Computational Modelling in Biology and Medicine
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. Takahashi, K., & Yamanaka, S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell, 126, 663-676.
[2]. Xu, R. L., et al. (2019). Correction of a pathogenic gene mutation in human embryos using CRISPR-Cas9.
[3]. Hochedlinger, K., & Plath, K. (2009). Epigenetic reprogramming and induced pluripotency. Development, 136, 509-523.
[4]. Wang, S. W., Gao, C., Zheng, Y. M., et al. (2022). Current applications and future perspective of CRISPR/Cas9 gene editing in cancer. Molecular Cancer, 21, 57.
[5]. Xiong, Z., & Jiang, Y. (2024). Research progress on methods to improve the efficacy of mesenchymal stem cell transplantation for treating liver cirrhosis. Journal of Gastroenterology and Hepatology, 33(10), 1401-1404.
[6]. Normile, D. (2017). China sprints ahead in CRISPR therapy race. Science, 358, 20-21.
[7]. Rupp, L. J., Schumann, K., Roybal, K. T., et al. (2017). CRISPR/Cas9-mediated PD-1 disruption enhances anti-tumor efficacy of human chimeric antigen receptor T cells. Scientific Reports, 7(1), 1-10.
[8]. Ou, X., Ma, Q., Yin, W., et al. (2021). CRISPR/Cas9 gene-editing in cancer immunotherapy: Promoting the present revolution in cancer therapy and exploring more. Frontiers in Cell and Developmental Biology, 9, 674467.
[9]. Feng, Y., Sassi, S., Shen, J. K., et al. (2015). Targeting Cdk11 in osteosarcoma cells using the CRISPR-Cas9 system. Journal of Orthopaedic Research, 33(2), 199-207.
[10]. Rath, J. A., & Arber, C. (2020). Engineering strategies to enhance TCR-based adoptive T cell therapy. Cells, 9(6), 1485.









