1. Introduction
1.1. Triple-Negative Breast Cancer
Triple-negative breast cancer (TNBC) lacks the expression of three key molecular targets: estrogen receptor (ER), progesterone receptor (PR), and HER2. It is one of the most common and deadly cancers in women [1]. Epidemiological surveys show that TNBC accounts for 15%–25% of new breast cancer cases. According to the 2020 Global Cancer Burden Data from the International Agency for Research on Cancer (IARC) and the World Health Organization (WHO), approximately 416,000 new breast cancer cases are diagnosed annually in China, with 60,000–80,000 involving TNBC [2]. Common risk factors for TNBC include family history, a high number of menstrual cycles, late pregnancy, and breastfeeding [3]. Comparing with other subtypes, TNBC has fewer treatment options. Chemotherapy remains the primary treatment, but the recurrence rate is high [3]. Compared to other breast cancer subtypes, TNBC has fewer treatment options. Currently, chemotherapy has been the primary treatment for TNBC patients, but the recurrence rate is extremely high [4]. While PARP inhibitors show targeted therapeutic potential, their effectiveness is limited to a subset of patients [5]. Overall, TNBC is highly aggressive, often diagnosed at advanced stages, and associated with poor prognosis and a high risk of visceral metastasis. Therefore, developing early diagnostic and treatment strategies is crucial for improving clinical outcomes.
1.2. Triple-Negative Breast Cancer Metastasis
Tumor metastasis leads to death of cancer patients, which involves not only changes in tumor cells but also changes in the tumor microenvironment [6]. Metastasis is the process by which tumor cells invade surrounding lymphatic and blood vessels, spread to other tissues, and form new tumor foci. Metastasis can be classified into four types: local spread, lymphatic metastasis, blood metastasis, and distant metastasis [7]. In 1889, Stephen Paget's "seed-soil" theory holds that cancer cell metastasis depends on the interaction between cancer cells ("seeds") and the specific organ microenvironment ("soil"). The tumor-directed metastasis theory indicates that certain tumors preferentially metastasize to specific organs [8]. TNBC is highly prone to early metastasis, with a median overall survival of only 18 months once metastasis occurs. Brain metastases (BM) are particularly life-threatening for TNBC patients [9]. Therefore, early diagnosis of TNBC patients at high risk of metastasis is crucial for timely treatment and prevention.
Biomarkers, which include proteins, RNA, and other molecules, are critical for disease diagnosis, especially for tumor metastasis. Biomarkers can detect early, low-level damage, offer early warnings, and provide diagnostic support [10]. Clinically, biomarkers are categorized as diagnostic markers (to confirm disease presence or subtypes), disease severity markers (to assess disease progression), and prognostic markers (to predict clinical events such as recurrence or metastasis) [11]. Thus, biomarkers are essential for diagnosing tumor metastasis.
The biomarkers for TNBC brain metastasis remain unclear. Yunzhu et al. analyzed tissue and cell line samples of TNBC brain metastasis and identified 15 differentially expressed genes. Their results suggest that CXCL8 could serve as a prognostic biomarker for brain metastases in TNBC. Treatment of MDA-MB-231 and Hs578t cell lines with recombinant CXCL8 further confirmed its role [12]. Although CXCL8 may be a potential marker, it has limitations. Tumors evolve rapidly, and detecting markers like CXCL8 often requires invasive biopsies. These procedures, while accurate, pose risks, especially for advanced-stage patients who cannot tolerate repeated procedures. Moreover, no clinical marker for TNBC brain metastasis currently exists, warranting further investigation into new biomarkers. Therefore, we aim to explore a sensitive, accurate, and less invasive biomarker for early TNBC brain metastasis diagnosis.
Circulating tumor cells (CTCs) are a promising foundation for a novel biomarker for distal metastasis [13]. CTCs originate from primary tumors and are considered key agents of metastasis. They can survive in the bloodstream, evade immune responses, and colonize distant organs [14]. The detection of CTCs offers a real-time liquid biopsy approach for TNBC brain metastasis, enabling early intervention, treatment decisions, and prognosis assessment [15]. In conclusion, peripheral blood markers may provide a sensitive, accurate, and minimally invasive method for the early diagnosis of TNBC brain metastasis in the future.
1.3. The Role of Noncoding RNA in Triple-Negative Breast Cancer Metastasis
Non-coding RNA (ncRNA) is an RNA molecule transcribed from the genome that does not encode proteins [16]. There are two main types: structural ncRNA and regulatory ncRNA. Regulatory ncRNA can be further divided into long non-coding RNA (lncRNA) and micro RNA (miRNA). LncRNAs are usually longer than 200 nucleotides and play important roles in epigenetic regulation, cell cycle control, and cell differentiation [17]. MiRNAs are small single-stranded molecules of 20-24 nucleotides that are derived from a hairpin structure called pre-miRNA. They regulate post-transcriptional gene expression by binding to complementary sequences in the 3' untranslated region (3'UTR) of target mRNAs, resulting in gene silencing [18]. Long intergenic noncoding RNAs (lincRNAs) can regulate gene expression by acting as competitive endogenous RNA (ceRNA) sponges, a mechanism that highlights RNA-RNA interactions [19]. LncRNAs make up a significant portion of human genome transcripts, accounting for up to 62% and existing in large numbers. Further research is needed to fully understand how functional lncRNAs influence tumor recurrence and metastasis, offering potential targets for cancer treatment [20].
LncRNAs play a pivotal role in TNBC metastasis. Research by Po-Shun et al. found that the lncRNA Linc-ZNF469-3 promotes TNBC lung metastasis through the miR-574-5p-ZEB1 signaling axis, potentially serving as a prognostic marker for TNBC metastasis [21]. Yongyin et al. observed elevated levels of LncRNA T376626 in TNBC serum and tissues, with cellular experiments showing that it binds to LAMC2, affecting TNBC cell invasion and migration. This suggests LncRNA T376626 as a diagnostic and prognostic biomarker for TNBC [22].LincRNAs also contribute to distant metastasis through circulating tumor cells (CTCs). Studies have identified DARS-AS1 as a potential therapeutic target for metastatic TNBC. Overexpression of DARS-AS1 promotes migration and invasion of human TNBC by activating the NF-κB/STAT3 pathway through inhibition of miR-129-2-3p and upregulation of CDK1. Treatment with DARS-AS1 siRNA-loaded exosomes effectively slows the growth and liver metastasis of TNBC [23]. The upregulation of lncRNA MLLT4-AS1 in TNBC tissues and Gln-deficient TNBC cell lines also promotes angiogenesis and metastasis by increasing Gln levels via XBP1SBM [24]. In conclusion, lincRNAs are crucial in TNBC metastasis, but their role in TNBC brain metastasis remains unclear and warrants further study.
1.4. Role of LINC02487 in Triple-Negative Breast Cancer Metastasis
Yanli et al. from Peking University People's Hospital conducted a study using 34 fresh breast tumor tissues obtained between June and December 2020 from patients diagnosed with primary breast cancer. These patients underwent surgical resection, and RNA was extracted from the tumor tissue. The cDNA library was sequenced using the NextSeq platform. A comparison of gene expression between TNBC tissue and other breast cancer subtypes identified 273 differentially expressed genes (DEGs) in TNBC, with 172 up-regulated and 101 down-regulated.
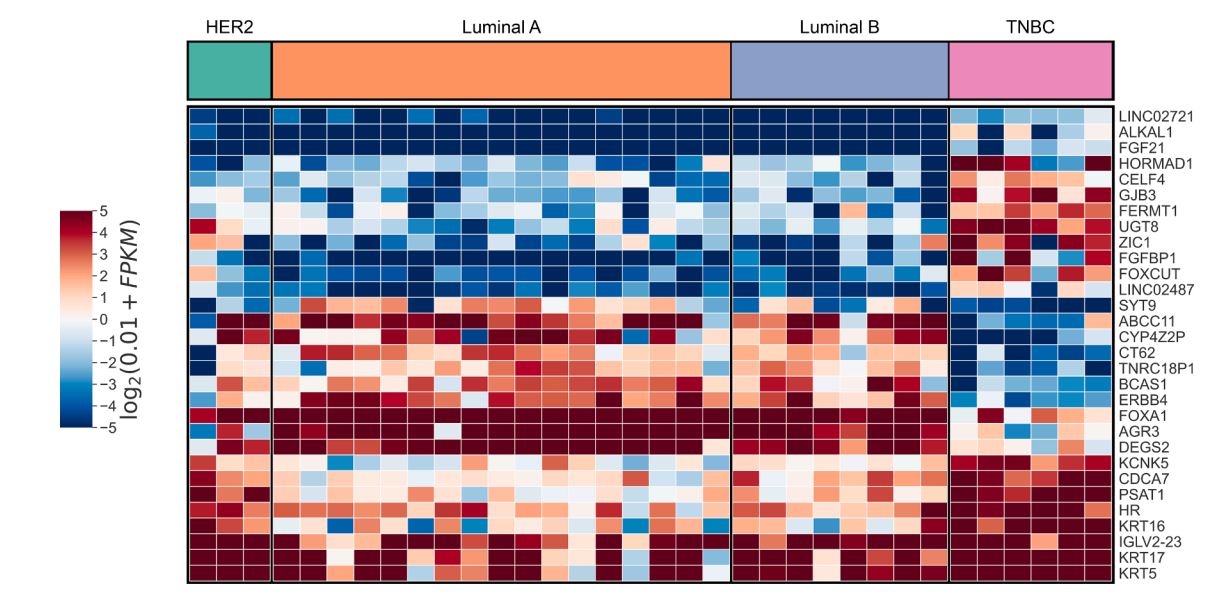
Figure 1: TNBC gene expression difference analysis heat map [25]
Notably, in Figure 1, LINC02487 was significantly up-regulated in TNBC, as shown in the heat map. However, its potential as an early warning marker for brain metastasis remains unclear [25]. Multiple studies suggest that LINC02487 could serve as an early diagnostic marker for various cancers. Joanna et al. found through bioinformatics analysis that LINC02487 may be a clinical molecular marker for head and neck squamous cell carcinoma [26]. Yue et al. analyzed clinical lncRNA data from 167 oral squamous cell carcinoma (OSCC) samples and 45 normal tissues, identifying six new lncRNA biomarkers for OSCC [27]. Feng et al. observed a significant reduction in LINC02487 expression in six OSCC cell lines. In a study of 50 OSCC samples from the Chinese population, they found a correlation between LINC02487 expression levels and cancer metastasis. Further research showed that LINC02487 interacts with USP17 (a deubiquitinating enzyme) and inhibits OSCC cell migration and invasion through the USP17-snai1 axis [28]. In summary, although LINC02487 shows potential as a marker for tumor metastasis, its role in TNBC metastasis remains undocumented.
In conclusion, LINC02487 is significantly up-regulated in the differential gene expression profile of TNBC. However, the specific role of LINC02487 in TNBC metastasis is still unknown, suggesting that it could become a promising clinical marker for TNBC metastasis in the future.
1.5. Research Significance
Brain metastasis is the main cause of death in TNBC patients, particularly impacting Chinese women. Currently, there exists no definitive prognostic marker for TNBC brain metastasis through liquid biopsy in clinical practice. This project, based on the gene expression profiles of TNBC patients at Peking University, introduces LINC02487 as a potential molecular marker for TNBC brain metastasis. Through cytological experiments, the molecular mechanism of LINC02487 will be further investigated using methods like RT-qPCR and RNA fluorescence detection. Successful completion of this project is anticipated to provide crucial scientific and clinical insights, potentially offering new molecular markers for TNBC brain metastasis.
2. Method
2.1. Patient Samples and Public Databases
The relative expression data of LINC02487 in triple-negative breast cancer (TNBC) patients were acquired from UALCAN, a publicly available cancer lncRNA interaction exploration resource (https://ualcan.path.uab.edu/). We adhered to the website guidelines and utilized datasets from The Cancer Genome Atlas (TCGA) for our analysis.
2.2. Cell Lines and Cell Culture
T-47D cell line (human breast ductal carcinoma cell line) and two triple-negative breast cancer (TNBC) cell lines BT-549 and MDA-MB-231 were obtained from Wuhan Pronocell Biotechnology Co., Ltd. The cell culture conditions and culture medium of this experiment were as follows: T-47D cells were cultured in RPMI 1640 medium; BT-549 cells were cultured in RPMI 1640 medium; MDA-MB-231 cells were cultured in DMEM medium. All culture media were supplemented with 10% (v/v) fetal bovine serum (Gibco) and 1% penicillin-streptomycin (100 μg/mL penicillin and 100 μg/mL streptomycin). After thawing, the cells were resuspended in the corresponding culture medium, centrifuged, and the supernatant was discarded. Fresh medium was added, and the cells were transferred to a culture dish, labeled, and placed in an incubator.
2.3. Lentiviral-Mediated Overexpression
In this study, the pCMV-MCS-PGK-Puro vector was used to achieve stable overexpression of LINC02487. The full-length LINC02487 sequence was synthesized and molecularly cloned into the pCMV-puro lentiviral vector (HanYin Biotech). The constructed overexpression vector and the empty pCMV-puro vector (as a control) were transfected into cells using lentivirus. To verify the overexpression efficiency of LINC02487, RT-qPCR was performed to measure its expression level.
2.4. Methods for Extracting Total RNA from Cells
Add 500 μl Trizol to each well of a 12-well plate and digest the cells on ice until completely lysed. Pipette the lysate repeatedly to ensure that the cells are completely broken. Transfer the mixture to a 1 ml centrifuge tube, incubate for 5-10 minutes, and then vortex with 500 μl chloroform for 15 seconds. Allow to separate at room temperature (the upper layer is chloroform) and transfer the supernatant to a new tube. Next, add 400 μl isopropanol and centrifuge for 10 minutes. Discard the supernatant and wash the RNA pellet with 1 ml 75% ethanol and centrifuge. Remove the ethanol and air-dry the pellet for 10-15 minutes. Finally, DEPC water was added to dissolve the RNA based on the amount of RNA precipitation.
2.5. RT-qPCR
Total cellular RNA was isolated, and cDNA was synthesized using the TB Green® Premix Ex Taq™ II Kit (Takara). Real-time qPCR was performed on an Applied Biosystems StepOnePlus Real-Time PCR System using the TB Green™ Premix Ex Taq™ PCR Kit (Takara Corporation) following the manufacturer’s instructions. The primer sequences used are shown in Table 1:
Table 1: Sequences of RT-qPCR primers
Positive | Reverse | |
Internal reference actin | GGGTATTTTGTGCTCCCCCA | CAGGCACTGAAGGTTCGGAT |
LINC02487 | GGGTATTTTGTGCTCCCCCA | CAGGCACTGAAGGTTCGGAT |
2.6. CCK-8
A total of 1,000 cells/well were seeded into 96-well plates. At 3 time points (24, 48, and 72 hours after seeding), 10 μL of CCK-8 reagent (Beyotime, C0039, Cell Counting Kit-8) and 100 μL of culture medium were added to each well and then incubated at 37 °C for 2 hours. The optical density (OD) at 450 nm was measured using a microplate reader. Data are from three independent experiments, each of which was repeated three times.
2.7. DNA Transfection
Before transfection, approximately 200,000 to 700,000 cells per well were seeded into a six-well plate and cultured until the cell density reached 70–90%. Then replace the medium in each well with fresh medium containing serum and no antibiotics. Prepare two clean sterile centrifuge tubes for each well of the six-well plate. Add 125 μl of DMEM medium containing antibiotics and serum to each tube. Next, add 2.5 μg of micro-DNA to one tube and mix gently by pipetting. Add 5 μl of Lipo6000™ transfection reagent to another tube and mix well. Then gently mix the two and incubate at room temperature for 5 minutes. Subsequently, add 250 μl of Lipo6000™-DNA mixture evenly to each well of the six-well plate and mix gently. After 24–48 hours of further incubation, the transfection efficiency was assessed using appropriate methods.
2.8. SiRNA Transfection
For each well of the six-well plate to be transfected, two centrifuge tubes were prepared. Add 125 μl of DMEM medium containing antibiotics and serum to each tube, add 100 pmol siRNA to one tube and mix thoroughly, and add 5 μl of Lipo6000™ transfection reagent to the other tube and mix thoroughly. The remaining steps were the same as for DNA transfection.
2.9. EdU Staining
EdU staining experiments were performed using the BeyoClick™ EdU-488 Cell Proliferation Assay Kit (C0071S, Beyotime). An appropriate amount of cells was cultured in a six-well plate and a 2×EdU working solution was prepared. An equal volume of preheated (37 °C) 2×EdU working solution (20 μM) was added to the six-well plate to ensure a final concentration of 1×EdU and incubated for 2 hours. After EdU labeling, the culture medium was removed, 1 ml of fixative was added, and the cells were fixed at a constant temperature for 15 minutes. The fixative was then removed and the cells were washed three times with 1 ml of wash solution for 3 to 5 minutes each time. Next, 1 ml of permeabilization solution was added and the cells were incubated at room temperature for 10 to 15 minutes.
2.10. Transwell Experiment
4 × 104 cells were suspended in 100 μL of DMEM without FBS and seeded into the upper chamber of a 24-well Transwell chamber (BD Pharmingen) with an 8 μm pore size membrane. The lower chamber was filled with medium containing 10% FBS as a chemoattractant. After 24 h of incubation, the non-migrated cells on the upper side of the membrane were removed with a cotton swab. The cells that migrated to the bottom of the membrane were fixed and stained with crystal violet solution (Beyotime Biotech), then counted and imaged under a microscope. Eight fields of view were randomly selected in each chamber to count the number of cells, and the average was calculated. Each experiment was repeated three times. The cell counting index was defined as the number of migrated cells.
2.11. Statistical Analysis
Statistical analysis was performed using GraphPad 7. The relative expression levels of RNA were analyzed using the 2−ΔΔCT method. All data are presented as the mean ± SD of three or more independent experiments. Two groups of data were compared using a two-sided Student's t test, with a significance level of P < 0.05.
3. Result
3.1. LINC02487 Being Significantly Upregulated in TNBC Cell Lines
RT-qPCR was performed to investigate the expression of LINC02487 in TNBC. The results in Figure 2A showed that LINC02487 was significantly upregulated in two TNBC cell lines, BT-549 (p < 0.001) and MDA-MB-231 (p < 0.0001), compared to the T-47D human breast ductal carcinoma cell line. The expression level in the MDA-MB-231 cell line was 10-fold higher than that in BT-549, prompting the selection of MDA-MB-231 for subsequent experiments. To validate these findings, the relative expression of LINC02487 in the TCGA data was analyzed (as shown in Figure 2B), and significantly higher expression was observed in 115 TNBC patients compared to controls, suggesting a potential association between LINC02487 expression and cancer proliferation and metastasis.
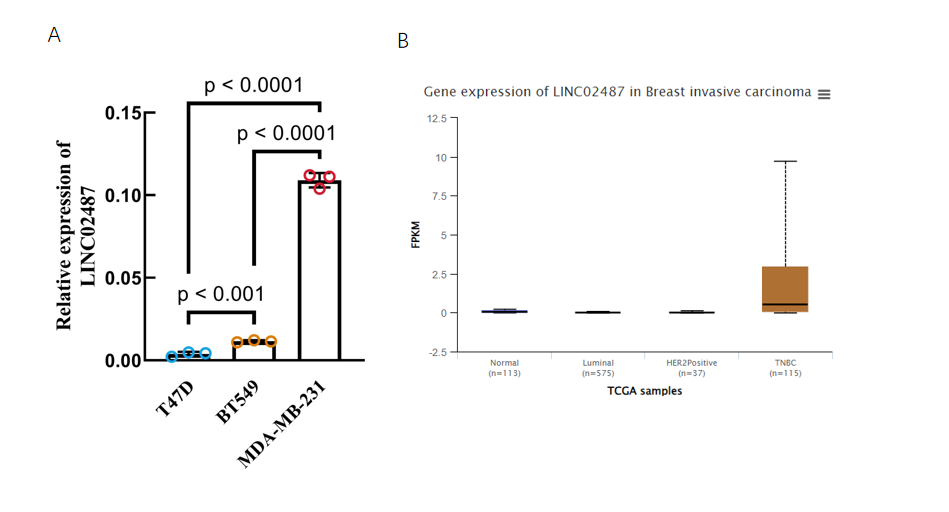
Figure 2: Expression level of LINC02487 in TNBC.
A | RT-qPCR was used to detect the expression level of LINC02487 in TNBC cell lines BT 549 and MDA-MB-231.
B | LINC02487 expression level of TNBC clinical samples in TCGA database.
3.2. LINC02487 Overexpression Promotes TNBC Cell Proliferation
To study the function of LINC02487 in TNBC, we established an MDA-MB-231 TNBC cell line overexpressing LINC02487 via lentiviral transfection of the pCMV-puro plasmid. RT-qPCR results showed that LINC02487 was stably overexpressed more than 3000 times after transfection into MDA-MB-231 cells as shown in Figure 2A. Next, we compared the proliferation phenotype changes of MDA-MB-231 cells in the LINC02487 overexpression group, empty vector group, and control group. The CCK-8 experiment found that the cell viability of LINC02487 overexpressing cells was significantly increased. Further testing was performed on cells from each group at three time points: 24 h, 48 h, and 72 h. The results showed that the empty vector overexpression group was significantly better than the control group. There was no significant difference in cell activity among the groups, but the activity of cells overexpressing LINC02487 was significantly higher than that of cells in the empty vector group, as shown in Figure 4A-C.
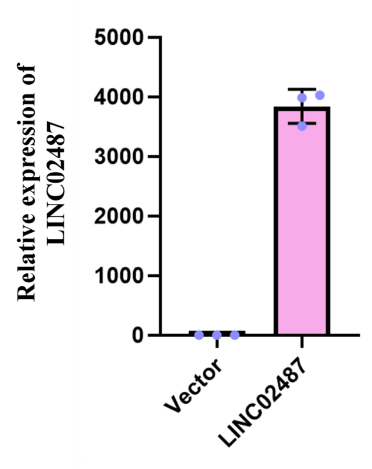
Figure 3: The relative expression level of LINC02487 after transfection with lentiviral plasmid overexpression vector
3.3. LINC02487 Knockdown Significantly Inhibits the Proliferation of TNBC Cells
To further confirm the function of LINC02487 in TNBC, we also examined the effect of knocking down LINC02487 on cell proliferation in MDA-MB-231 cells. The CCK-8 test found that the cell viability of cells after LINC02487 knockdown was significantly reduced, as observed in Figure 4A-C. Among them, cells were taken for testing at three time points: 24 h, 48 h, and 72 h. The results showed that the activity of LINC02487 knockdown cells significantly decreased over time, and the difference with the control group was most obvious at 48 h. Statistical analysis in Figure 4 at 48 h revealed that cell activity in the LINC02487 knockdown group was significantly inhibited compared to the control group (P < 0.0001), and was also significantly lower than in the empty vector control group (P < 0.0001).
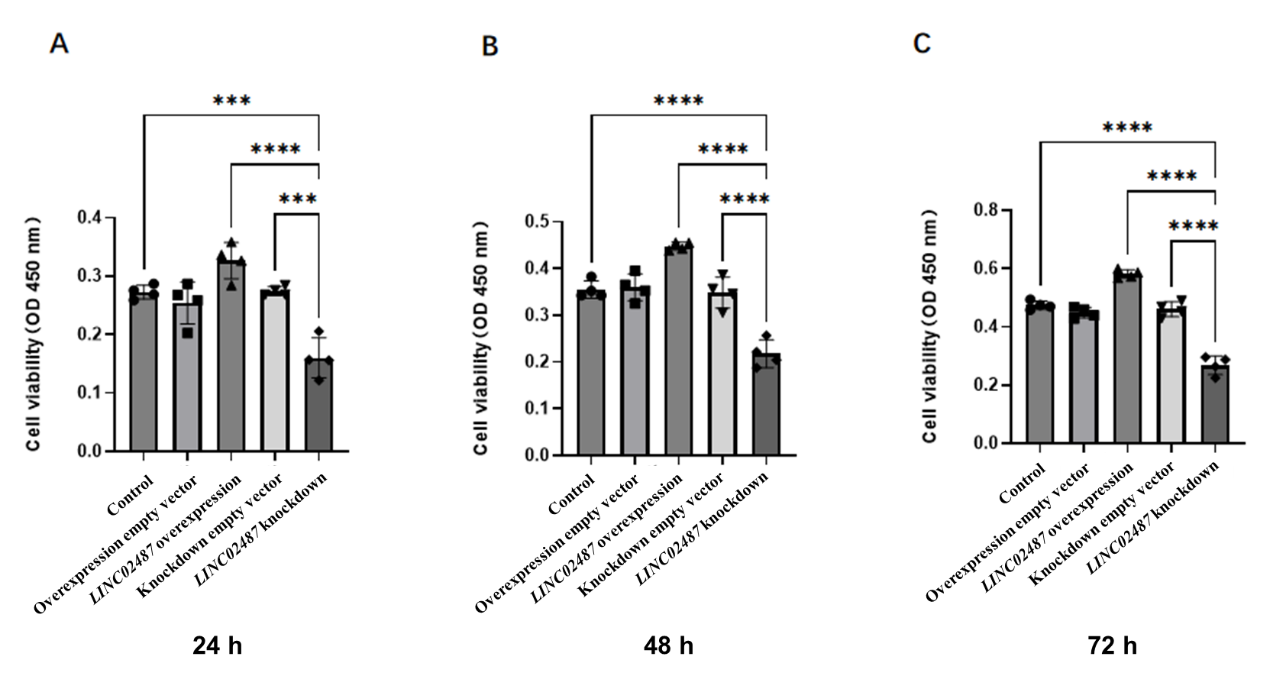
Figure 4: Effects of LINC02487 overexpression and knockdown on MDA-MB-231 Effects on cell proliferation
A-C | CCK-8 assay detected the changes in cell proliferation at different time points of 24 h, 48 h, and 72 h after LINC02487 overexpression or knockdown. **P < 0.01, ***P < 0.001, ****P < 0.0001.
3.4. EDU staining to detect whether LINC02487 affects TNBC cell viability
EDU (5-ethynyl-2'-deoxyuridine) is a novel thymidine analog that can replace thymidine and is incorporated into newly synthesized DNA during DNA replication. It is commonly used as a marker to detect cell activity. To further confirm the effect of LINC02487 on TNBC cell activity, we stained four groups of cells—overexpressing empty vector, overexpressing LINC02487, knocking down empty vector, and knocking down LINC02487—with EDU to assess cell activity. The results in Figure 5 showed that, compared to the empty vector control group, the cell activity in the LINC02487 overexpression group was the highest, while the LINC02487 knockdown group exhibited the lowest activity. What’s more, we used a double staining method, marking cells with HOECHST nuclear staining to detect apoptosis and clarify the results.
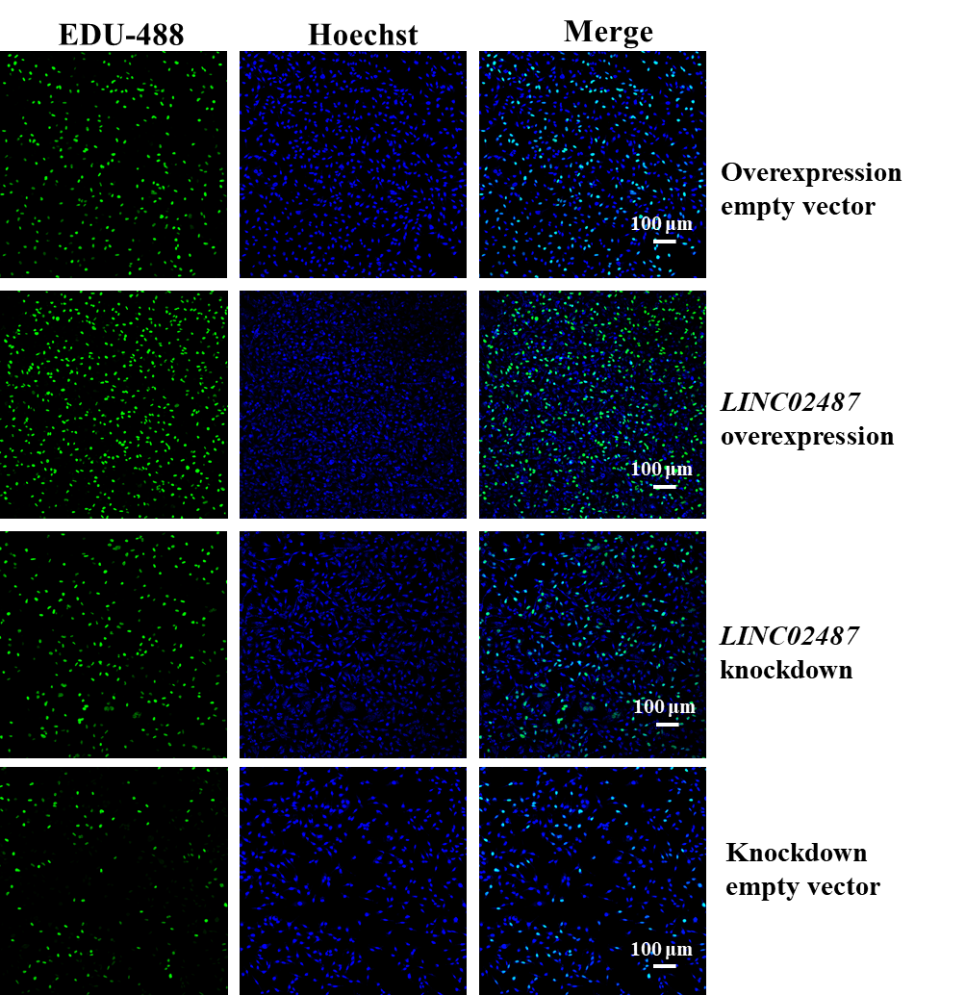
Figure 5: EDU staining to detect the effect of LINC02487 on TNBC cell viability
The four groups of cells in the figure are LINC02487 knockdown, LINC02487 overexpression, knockdown empty vector, and overexpression empty vector; green fluorescence represents EDU staining; blue fluorescence represents HOECHST staining
3.5. Transwell Assay to Detect Whether LINC02487 Affects the Migration and Invasion of TNBC Cells
We examined the effect on the migration and invasion abilities of MDA-MB-231 cells to further explore the function of LINC02487. We performed Transwell experiments, with or without Matrigel-coated upper chambers, on four groups of cells. The results in Figure 6A showed that both migration and invasion were significantly inhibited in LINC02487 knockdown cells. Comparison of the LINC02487 overexpression group with the empty vector overexpression group revealed a significant increase in both migration and invasion of TNBC cells. Similarly, comparing the LINC02487 knockdown group with the empty vector knockdown group in Figure 6A and B showed a significant improvement in migration and invasion, with values significantly higher than those in the LINC02487 overexpression group.
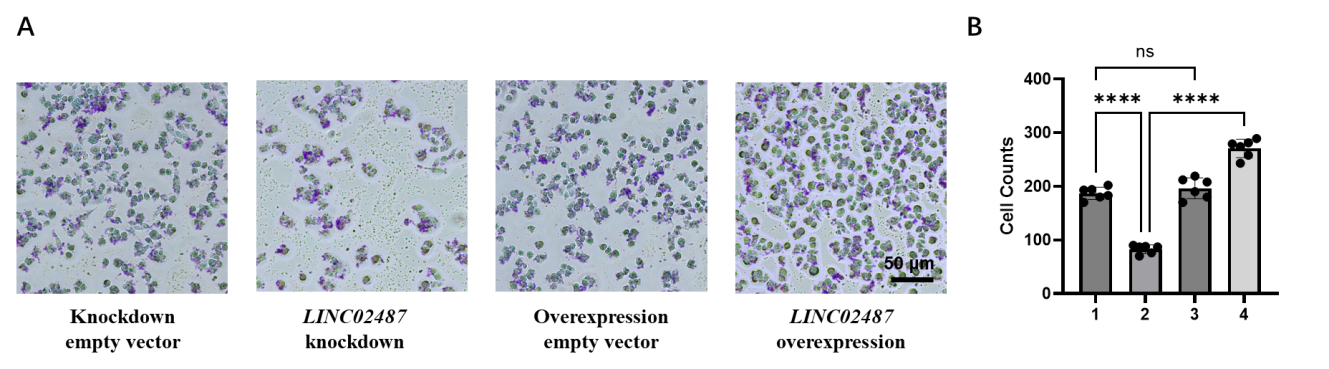
Figure 6: Transwell assay detects that LINC02487 affects the migration and invasion of TNBC cells.
A | Using Transwell assay to detect cell migration in LINC02487 knockdown, LINC02487 overexpression, knockdown empty vector, and overexpression empty vector.
B | Statistical data of cell numbers in the above four groups. ns, not significant; *P < 0.05, **P < 0.01, ***P < 0.001.
4. Discussion
In this study, based on second-generation sequencing results from five TNBC patients published by Peking University People’s Hospital, we discovered that the expression of two long non-coding RNAs, LINC02487 and LINC02721, was specifically upregulated in TNBC. We first demonstrated that the expression of LINC02487 was significantly upregulated in both TNBC cell lines and in 115 TNBC patients from the TCGA public database. To further explore the involvement of LINC02487 in regulating the pathogenesis of TNBC, particularly in proliferation and distant metastasis, we used genetic engineering methods to construct in vitro models of TNBC with LINC02487 overexpression and knockdown. CCK-8 assays showed that LINC02487 overexpression significantly increased proliferation in the MDA-MB-231 cell line, while LINC02487 knockdown significantly inhibited proliferation and delayed TNBC progression. Finally, we confirmed that LINC02487 significantly influences the proliferation and metastasis of the MDA-MB-231 TNBC cell line. EDU staining results further validated the CCK-8 findings at the cellular level, showing that knockdown of LINC02487 significantly inhibited proliferation in MDA-MB-231 cells. Transwell assays confirmed that LINC02487 knockdown inhibited the migration and invasion of TNBC cells, delaying disease progression, while LINC02487 overexpression promoted migration and invasion, increasing the risk of distant metastasis [29]. To explore the function of LINC02487 in TNBC, we used StarBase to predict its target genes and, based on existing literature, speculated on the potential molecular mechanisms through which LINC02487 promotes proliferation and metastasis in TNBC. Taken together, these findings suggest that LINC02487 may be a potential prognostic biomarker for TNBC progression and metastasis, playing a novel role in TNBC pathogenesis.
LncRNA expression can serve as a marker for identifying biomarkers related to the prognosis of TNBC and is crucial for improving the biological understanding of the mechanisms driving TNBC progression [29]. LncRNAs may function as epigenetic, transcriptional, and post-transcriptional regulators in various cancer types [30]. The results show that the expression level of LINC02487, compared to the T47D human breast duct carcinoma cell line, is significantly upregulated in two TNBC cell lines (BT-549 and MDA-MB-231), with a more prominent upregulation observed in MDA-MB-231 cells. Yanli et al. from Peking University People’s Hospital collected clinical samples from five TNBC patients and found, through second-generation sequencing, that the expression of LINC02487 was significantly upregulated in Chinese TNBC patients, which is consistent with our findings [25]. In Yu's study, it was also found that LINC02487 and FOXCUT are specifically highly expressed in basal-like breast cancer tissues. The increased expression of LINC02487 may be positively correlated with a poorer prognosis. Their study further supports the idea that LINC02487 may be highly expressed in TNBC [31]. Although our data shows that LINC02487 is highly expressed in TNBC, studies have also reported that LINC02487 is poorly expressed in oral squamous cell carcinoma (OSCC) and cervical cancer (CC), which contrasts with our results. Feng et al. confirmed that the expression level of LINC02487 was significantly reduced in six independent OSCC cell lines. Further studies found that LINC02487 expression was significantly downregulated compared to paired adjacent normal tissues in 50 OSCC samples from the Chinese population, and this may be related to tumor metastasis [27, 28]. Ru et al.'s study found that, similar to OSCC, LINC02487 expression was downregulated in CC cells and tissues, and it may inhibit the progression of CC by upregulating PTEN expression and inhibiting the Akt/mTOR signaling pathway [32].
We found that the expression level of LINC02487 is inconsistent with its expression trend in OSCC and CC, suggesting that during tumor proliferation and metastasis, the mode of action of LINC02487 differs significantly between TNBC, OSCC, and CC. This difference may be due to distinct targets and pathways involved in each cancer type. We also analyzed the expression of LINC02487 across various cancer types in the TCGA database, as shown in Figure 7. Compared to OSCC samples, LINC02487 expression was significantly upregulated in normal tissues. Additionally, figure 7 shows that the expression level of LINC02487 was significantly upregulated in colorectal cancer, cholangiocarcinoma, rectal cancer, and esophageal cancer samples. Thus, the trend of LINC02487 expression may vary across different cancer types.
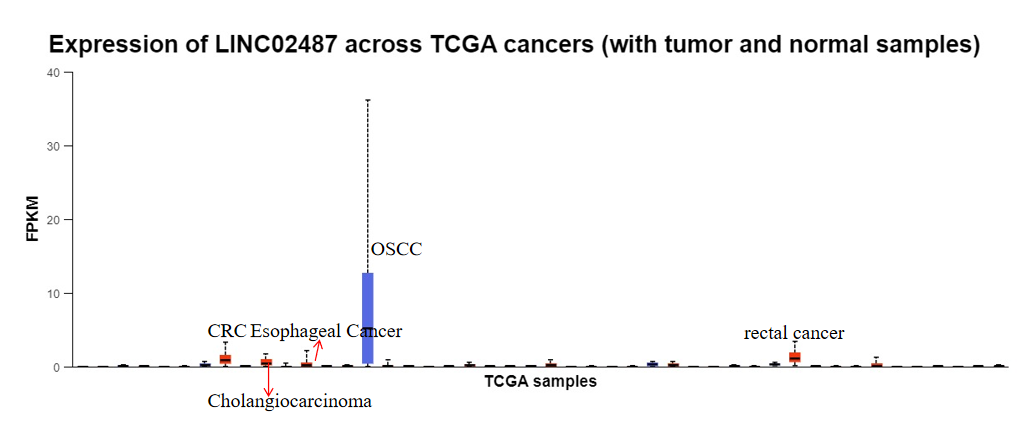
Figure 7: Expression levels of LINC02487 in pan-cancers in TCGC public data
LncRNA expression has important implications for the transcriptome and is dysregulated in many cancer types, and often serves as a prognostic marker. Upregulation of LINC02487 may act as a prognostic marker for TNBC metastasis. To verify this hypothesis, we examined the relationship between high LINC02487 expression in breast cancer and overall survival (OS) in TCGA data, as being presented in Figure 8. Our findings show that the OS of breast cancer patients with high LINC02487 expression is not significantly reduced. While high LINC02487 expression in breast cancer does not lower overall survival, it does not rule out a potential link with TNBC patient survival. Future studies could explore the correlation between high LINC02487 expression and survival rates in TNBC patients, providing further evidence for its use as a prognostic marker.
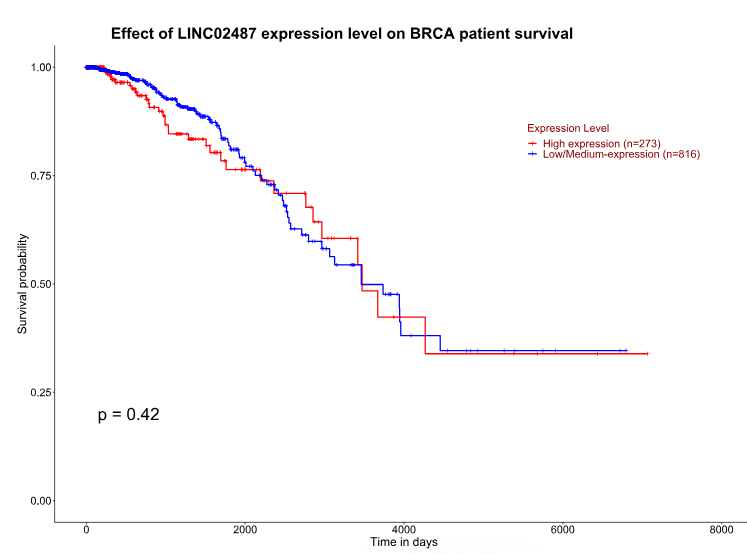
Figure 8: Correlation between high expression of LINC02487 and OS of breast cancer patients in TCGC public data
The StarBase v2.0 database is used to predict the potential downstream target genes of LINC02487 in TNBC, being shown in Figure 9. Our analysis in Figure 9 identified AIF1L (Allograft Inflammatory Factor 1 Like), PCGF2 (Polycomb Group RING Finger Protein 2), and MIAT (Myocardial Infarction Associated Transcript) as potential targets. According to Pei et al., AIF1L is downregulated and hypermethylated in breast cancer patients, and its ectopic expression inhibits MDA-MB-231 cell migration and invasion. Further evidence showed that AIF1L overexpression inhibited cell spreading, altered cell shape, and reduced protrusion formation because of decreased expression of focal adhesion kinase (FAK) and RhoA [33]. A cohort study from Shanghai found altered expression of PCGF2 in TNBC patients, though no further mechanistic studies were conducted [34]. PCGF2, a transcriptional repressor often located in the nucleus, participates in DNA-binding transcription factor activity and ubiquitin protein transferase activity. It has tumor suppressor activity and may regulate cancer cell proliferation. Studies have shown that LINC02487 is located in the cytoplasm and accumulates near the nuclear membrane [28]. Therefore, we hypothesize that LINC02487 may regulate TNBC-related pathogenic gene transcription in the nucleus through PCGF2. The expression of MIAT is significantly higher in TNBC compared to normal or adjacent tissues, which may be linked to tumor-infiltrating immune cells in the tumor microenvironment [35]. Future experiments will further verify the binding and functionality of LINC02487 with AIF1L, PCGF2, and MIAT through ChIRP, WB, and RIP assays. Based on this bioinformatics analysis combined with literature evidence, we conclude that the cellular functions regulated by LINC02487 may be mediated through PCGF2.
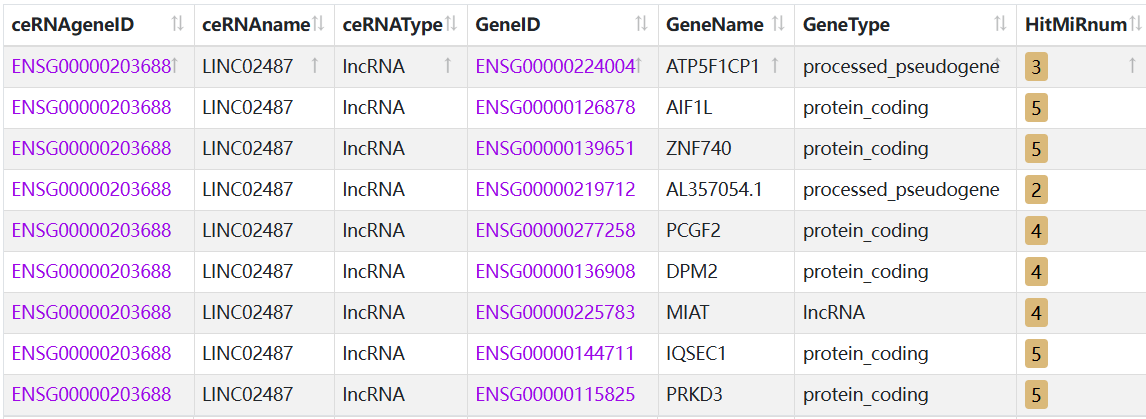
Figure 9: Target genes of LINC02487 predicted by starbase v2.0 database
Table 2: Functions of potential target genes of LINC02487 in TNBC
Gene | Function | Signaling pathways in TNBC | |
AIF1L | Regulates actin filament binding activity and participates in cytoskeletal movement | Ectopic expression of AIF1L inhibited the migration and invasion of MDA-MB-231 cells, confirming that AIF1L overexpression inhibited cell spreading, changed cell shape, and reduced protrusion formation, which was associated with decreased expression of focal adhesion kinase (FAK) and RhoA. | [33] |
PCGF2 | Transcription repressor. DNA-binding transcription factor activity and ubiquitin-protein transferase activity. May inhibit tumor activity. Plays a role in controlling cancer cell proliferation. | PCGF2 specifically binds to the DNA sequence 5'-GACTNGACT-3'. Participates in the p38 MAPK signaling pathway | [34] |
MIAT | Encoding spliced long noncoding RNAs that may constitute components of the nuclear matrix. | unknown | [35] |
Due to limited resources, this experiment only involved cell-based assays to verify our hypothesis, with no animal experiments or clinical trials conducted. As the study was primarily in vitro, the results may differ in a human environment. In the future, we plan to conduct animal experiments to enhance the project. By constructing a TNBC animal model and testing drug efficacy on cancer tissue, we aim to gather in vivo data.
5. Conclusion
We used genetic engineering to construct in vitro TNBC models with LINC02487 overexpression and knockdown. CCK-8 assays showed that LINC02487 overexpression significantly increased the proliferation of the MDA-MB-231 cell line, whereas LINC02487 knockdown significantly inhibited its proliferation and delayed TNBC progression. We also determined that LINC02487 significantly affects the proliferation and metastasis of MDA-MB-231 cells. EDU staining further validated the CCK-8 results at the cellular level, confirming that LINC02487 knockdown significantly inhibited MDA-MB-231 proliferation. Transwell assays showed that LINC02487 knockdown inhibited migration and invasion of TNBC cells, while overexpression promoted these processes. Using StarBase, we predicted the target genes of LINC02487 and, in combination with literature, speculated on the potential molecular mechanisms underlying its role in TNBC proliferation and metastasis. In summary, LINC02487 may serve as a potential prognostic biomarker for TNBC progression and metastasis and play a novel role in its pathogenesis.
References
[1]. S. Loibl, P. Poortmans, M. Morrow, C. Denkert, and G. Curigliano, "Breast cancer," , Lancet (London, England), vol. 397, no. 10286, pp. 1750-1769, May 8 2021, doi: 10.1016/s0140-6736(20)32381-3.
[2]. M. Arnedos, C. Bihan, S. Delaloge, and F. Andre, "Triple-negative breast cancer: are we making headway at least?", Therapeutic advances in medical oncology, vol. 4, no. 4, pp. 195-210, Jul 2012, doi: 10.1177/1758834012444711.
[3]. A. M. Brewster, M. Chavez-MacGregor, and P. Brown, "Epidemiology, biology, and treatment of triple-negative breast cancer in women of African ancestry," , The Lancet. Oncology, vol. 15, no. 13, pp. e625-e634, Dec 2014, doi: 10.1016/s1470-2045(14)70364-x.
[4]. X. Bai, J. Ni, J. Beretov, P. Graham, and Y. Li, "Triple-negative breast cancer therapeutic resistance: Where is the Achilles' heel?," , Cancer letters, vol. 497, pp. 100-111, Jan 28 2021, doi: 10.1016/j.canlet.2020.10.016.
[5]. A. K. Mehta et al., "Targeting immunosuppressive macrophages overcomes PARP inhibitor resistance in BRCA1-associated triple-negative breast cancer," , Nature cancer, vol. 2, no. 1, pp. 66-82, Jan 2021, doi: 10.1038/s43018-020-00148-7.
[6]. Y. Lin, J. Xu, and H. Lan, "Tumor-associated macrophages in tumor metastasis: biological roles and clinical therapeutic applications," , Journal of hematology & oncology, vol. 12, no. 1, p. 76, Jul 12 2019, doi: 10.1186/s13045-019-0760-3.
[7]. Z. Meng, R. Zhang, X. Wu, M. Zhang, and T. Jin, "PD‑L1 mediates triple‑negative breast cancer evolution via the regulation of TAM/M2 polarization," , International journal of oncology, vol. 61, no. 6, Dec 2022, doi: 10.3892/ijo.2022.5440.
[8]. I. J. Fidler and G. Poste, "The "seed and soil" hypothesis revisited," , The Lancet. Oncology, vol. 9, no. 8, p. 808, Aug 2008, doi: 10.1016/s1470-2045(08)70201-8.
[9]. Y. Lv, X. Ma, Y. Du, and J. Feng, "Understanding Patterns of Brain Metastasis in Triple-Negative Breast Cancer and Exploring Potential Therapeutic Targets," , OncoTargets and therapy, vol. 14, pp. 589-607, 2021, doi: 10.2147/ott.s293685.
[10]. J. Sukumar, K. Gast, D. Quiroga, M. Lustberg, and N. Williams, "Triple-negative breast cancer: promising prognostic biomarkers currently in development," , Expert review of anticancer therapy, vol. 21, no. 2, pp. 135-148, Feb 2021, doi: 10.1080/14737140.2021.1840984.
[11]. J. K. Aronson and R. E. Ferner, "Biomarkers-A General Review," , Current protocols in pharmacology, vol. 76, pp. 9.23.1-9.23.17, Mar 17 2017, doi: 10.1002/cpph.19.
[12]. Y. Shen, B. Zhang, X. Wei, X. Guan, and W. Zhang, "CXCL8 is a prognostic biomarker and correlated with TNBC brain metastasis and immune infiltration," , International immunopharmacology, vol. 103, p. 108454, Feb 2022, doi: 10.1016/j.intimp.2021.108454.
[13]. M. J. Magbanua et al., "Circulating tumor cell analysis in metastatic triple-negative breast cancers," , Clinical cancer research : an official journal of the American Association for Cancer Research, vol. 21, no. 5, pp. 1098-105, Mar 1 2015, doi: 10.1158/1078-0432.ccr-14-1948.
[14]. F. C. Bidard, C. Proudhon, and J. Y. Pierga, "Circulating tumor cells in breast cancer," , Molecular oncology, vol. 10, no. 3, pp. 418-30, Mar 2016, doi: 10.1016/j.molonc.2016.01.001.
[15]. C. Riebensahm et al., "Clonality of circulating tumor cells in breast cancer brain metastasis patients," , Breast cancer research : BCR, vol. 21, no. 1, p. 101, Sep 3 2019, doi: 10.1186/s13058-019-1184-2.
[16]. H. Yan and P. Bu, "Non-coding RNA in cancer," , Essays in biochemistry, vol. 65, no. 4, pp. 625-639, Oct 27 2021, doi: 10.1042/ebc20200032.
[17]. M. C. Bridges, A. C. Daulagala, and A. Kourtidis, "LNCcation: lncRNA localization and function," , The Journal of cell biology, vol. 220, no. 2, Feb 1 2021, doi: 10.1083/jcb.202009045.
[18]. M. Hill and N. Tran, "miRNA interplay: mechanisms and consequences in cancer," , Disease models & mechanisms, vol. 14, no. 4, Apr 1 2021, doi: 10.1242/dmm.047662.
[19]. M. Cesana et al., "A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA," , Cell, vol. 147, no. 2, pp. 358-69, Oct 14 2011, doi: 10.1016/j.cell.2011.09.028.
[20]. S. J. Liu, H. X. Dang, D. A. Lim, F. Y. Feng, and C. A. Maher, "Long noncoding RNAs in cancer metastasis," , Nature reviews. Cancer, vol. 21, no. 7, pp. 446-460, Jul 2021, doi: 10.1038/s41568-021-00353-1.
[21]. P. S. Wang et al., "A novel long non-coding RNA linc-ZNF469-3 promotes lung metastasis through miR-574-5p-ZEB1 axis in triple negative breast cancer," , Oncogene, vol. 37, no. 34, pp. 4662-4678, Aug 2018, doi: 10.1038/s41388-018-0293-1.
[22]. Y. He et al., "LncRNA T376626 is a promising serum biomarker and promotes proliferation, migration, and invasion via binding to LAMC2 in triple-negative breast cancer," , Gene, vol. 860, p. 147227, Apr 15 2023, doi: 10.1016/j.gene.2023.147227.
[23]. X. Liu et al., "Exosomes deliver lncRNA DARS-AS1 siRNA to inhibit chronic unpredictable mild stress-induced TNBC metastasis," , Cancer letters, vol. 543, p. 215781, Sep 1 2022, doi: 10.1016/j.canlet.2022.215781.
[24]. X. Liu et al., "Exosomes deliver lncRNA DARS-AS1 siRNA to inhibit chronic unpredictable mild stress-induced TNBC metastasis," , Cancer letters, vol. 543, p. 215781, Sep 1 2022, doi: 10.1016/j.canlet.2022.215781.
[25]. Y. L. Chen et al., "Novel biomarkers identified in triple-negative breast cancer through RNA-sequencing," , Clinica chimica acta; international journal of clinical chemistry, vol. 531, pp. 302-308, Jun 1 2022, doi: 10.1016/j.cca.2022.04.990.
[26]. J. Kozłowska et al., "Long Intergenic Non-Coding RNAs in HNSCC: From "Junk DNA" to Important Prognostic Factor," , Cancers, vol. 13, no. 12, Jun 12 2021, doi: 10.3390/cancers13122949.
[27]. Y. Li, X. Cao, and H. Li, "Identification and Validation of Novel Long Non-coding RNA Biomarkers for Early Diagnosis of Oral Squamous Cell Carcinoma," , Frontiers in bioengineering and biotechnology, vol. 8, p. 256, 2020, doi: 10.3389/fbioe.2020.00256.
[28]. L. Feng, J. Zhang, M. Sun, F. Qiu, W. Chen, and W. Qiu, "Tumor Suppressor LINC02487 Inhibits Oral Squamous Cell Carcinoma Cell Migration and Invasion Through the USP17-SNAI1 Axis," , Frontiers in oncology, vol. 10, p. 559808, 2020, doi: 10.3389/fonc.2020.559808.
[29]. S. R. Volovat et al., "MiRNA and LncRNA as Potential Biomarkers in Triple-Negative Breast Cancer: A Review," , Frontiers in oncology, vol. 10, p. 526850, 2020, doi: 10.3389/fonc.2020.526850.
[30]. J. S. Mattick et al., "Long non-coding RNAs: definitions, functions, challenges and recommendations," , Nature reviews. Molecular cell biology, vol. 24, no. 6, pp. 430-447, Jun 2023, doi: 10.1038/s41580-022-00566-8.
[31]. X. Yu et al., "Promotion effect of FOXCUT as a microRNA sponge for miR-24-3p on progression in triple-negative breast cancer through the p38 MAPK signaling pathway," , Chinese medical journal, vol. 137, no. 1, pp. 105-114, Jan 5 2024, doi: 10.1097/cm9.0000000000002700.
[32]. R. Sun, S. L. He, H. Y. Liu, X. W. Wang, and S. H. Li, "Tumor Suppressor LINC02487 Inhibits the Progression of Cervical Cancer in Vitro by Regulating the PTEN/Akt/mTOR Pathway," , Discovery medicine, vol. 36, no. 187, pp. 1732-1742, Aug 2024, doi: 10.24976/Discov.Med.202436187.159.
[33]. P. Liu, W. Li, Y. Hu, and Y. Jiang, "Absence of AIF1L contributes to cell migration and a poor prognosis of breast cancer," , OncoTargets and therapy, vol. 11, pp. 5485-5498, 2018, doi: 10.2147/ott.s165874.
[34]. S. Wang et al., "Gene expression in triple-negative breast cancer in relation to survival," , Breast cancer research and treatment, vol. 171, no. 1, pp. 199-207, Aug 2018, doi: 10.1007/s10549-018-4816-9.
[35]. T. Ye et al., "LncRNA MIAT Services as a Noninvasive Biomarker for Diagnosis and Correlated with Immune Infiltrates in Breast Cancer," , International journal of women's health, vol. 13, pp. 991-1004, 2021, doi: 10.2147/ijwh.s312714.
Cite this article
Guo,Y. (2025). Long Noncoding RNA LINC02487 in the Proliferation and Metastasis of Triple-Negative Breast Cancer Cells. Theoretical and Natural Science,99,70-84.
Data availability
The datasets used and/or analyzed during the current study will be available from the authors upon reasonable request.
Disclaimer/Publisher's Note
The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of EWA Publishing and/or the editor(s). EWA Publishing and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
About volume
Volume title: Proceedings of the 5th International Conference on Biological Engineering and Medical Science
© 2024 by the author(s). Licensee EWA Publishing, Oxford, UK. This article is an open access article distributed under the terms and
conditions of the Creative Commons Attribution (CC BY) license. Authors who
publish this series agree to the following terms:
1. Authors retain copyright and grant the series right of first publication with the work simultaneously licensed under a Creative Commons
Attribution License that allows others to share the work with an acknowledgment of the work's authorship and initial publication in this
series.
2. Authors are able to enter into separate, additional contractual arrangements for the non-exclusive distribution of the series's published
version of the work (e.g., post it to an institutional repository or publish it in a book), with an acknowledgment of its initial
publication in this series.
3. Authors are permitted and encouraged to post their work online (e.g., in institutional repositories or on their website) prior to and
during the submission process, as it can lead to productive exchanges, as well as earlier and greater citation of published work (See
Open access policy for details).
References
[1]. S. Loibl, P. Poortmans, M. Morrow, C. Denkert, and G. Curigliano, "Breast cancer," , Lancet (London, England), vol. 397, no. 10286, pp. 1750-1769, May 8 2021, doi: 10.1016/s0140-6736(20)32381-3.
[2]. M. Arnedos, C. Bihan, S. Delaloge, and F. Andre, "Triple-negative breast cancer: are we making headway at least?", Therapeutic advances in medical oncology, vol. 4, no. 4, pp. 195-210, Jul 2012, doi: 10.1177/1758834012444711.
[3]. A. M. Brewster, M. Chavez-MacGregor, and P. Brown, "Epidemiology, biology, and treatment of triple-negative breast cancer in women of African ancestry," , The Lancet. Oncology, vol. 15, no. 13, pp. e625-e634, Dec 2014, doi: 10.1016/s1470-2045(14)70364-x.
[4]. X. Bai, J. Ni, J. Beretov, P. Graham, and Y. Li, "Triple-negative breast cancer therapeutic resistance: Where is the Achilles' heel?," , Cancer letters, vol. 497, pp. 100-111, Jan 28 2021, doi: 10.1016/j.canlet.2020.10.016.
[5]. A. K. Mehta et al., "Targeting immunosuppressive macrophages overcomes PARP inhibitor resistance in BRCA1-associated triple-negative breast cancer," , Nature cancer, vol. 2, no. 1, pp. 66-82, Jan 2021, doi: 10.1038/s43018-020-00148-7.
[6]. Y. Lin, J. Xu, and H. Lan, "Tumor-associated macrophages in tumor metastasis: biological roles and clinical therapeutic applications," , Journal of hematology & oncology, vol. 12, no. 1, p. 76, Jul 12 2019, doi: 10.1186/s13045-019-0760-3.
[7]. Z. Meng, R. Zhang, X. Wu, M. Zhang, and T. Jin, "PD‑L1 mediates triple‑negative breast cancer evolution via the regulation of TAM/M2 polarization," , International journal of oncology, vol. 61, no. 6, Dec 2022, doi: 10.3892/ijo.2022.5440.
[8]. I. J. Fidler and G. Poste, "The "seed and soil" hypothesis revisited," , The Lancet. Oncology, vol. 9, no. 8, p. 808, Aug 2008, doi: 10.1016/s1470-2045(08)70201-8.
[9]. Y. Lv, X. Ma, Y. Du, and J. Feng, "Understanding Patterns of Brain Metastasis in Triple-Negative Breast Cancer and Exploring Potential Therapeutic Targets," , OncoTargets and therapy, vol. 14, pp. 589-607, 2021, doi: 10.2147/ott.s293685.
[10]. J. Sukumar, K. Gast, D. Quiroga, M. Lustberg, and N. Williams, "Triple-negative breast cancer: promising prognostic biomarkers currently in development," , Expert review of anticancer therapy, vol. 21, no. 2, pp. 135-148, Feb 2021, doi: 10.1080/14737140.2021.1840984.
[11]. J. K. Aronson and R. E. Ferner, "Biomarkers-A General Review," , Current protocols in pharmacology, vol. 76, pp. 9.23.1-9.23.17, Mar 17 2017, doi: 10.1002/cpph.19.
[12]. Y. Shen, B. Zhang, X. Wei, X. Guan, and W. Zhang, "CXCL8 is a prognostic biomarker and correlated with TNBC brain metastasis and immune infiltration," , International immunopharmacology, vol. 103, p. 108454, Feb 2022, doi: 10.1016/j.intimp.2021.108454.
[13]. M. J. Magbanua et al., "Circulating tumor cell analysis in metastatic triple-negative breast cancers," , Clinical cancer research : an official journal of the American Association for Cancer Research, vol. 21, no. 5, pp. 1098-105, Mar 1 2015, doi: 10.1158/1078-0432.ccr-14-1948.
[14]. F. C. Bidard, C. Proudhon, and J. Y. Pierga, "Circulating tumor cells in breast cancer," , Molecular oncology, vol. 10, no. 3, pp. 418-30, Mar 2016, doi: 10.1016/j.molonc.2016.01.001.
[15]. C. Riebensahm et al., "Clonality of circulating tumor cells in breast cancer brain metastasis patients," , Breast cancer research : BCR, vol. 21, no. 1, p. 101, Sep 3 2019, doi: 10.1186/s13058-019-1184-2.
[16]. H. Yan and P. Bu, "Non-coding RNA in cancer," , Essays in biochemistry, vol. 65, no. 4, pp. 625-639, Oct 27 2021, doi: 10.1042/ebc20200032.
[17]. M. C. Bridges, A. C. Daulagala, and A. Kourtidis, "LNCcation: lncRNA localization and function," , The Journal of cell biology, vol. 220, no. 2, Feb 1 2021, doi: 10.1083/jcb.202009045.
[18]. M. Hill and N. Tran, "miRNA interplay: mechanisms and consequences in cancer," , Disease models & mechanisms, vol. 14, no. 4, Apr 1 2021, doi: 10.1242/dmm.047662.
[19]. M. Cesana et al., "A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA," , Cell, vol. 147, no. 2, pp. 358-69, Oct 14 2011, doi: 10.1016/j.cell.2011.09.028.
[20]. S. J. Liu, H. X. Dang, D. A. Lim, F. Y. Feng, and C. A. Maher, "Long noncoding RNAs in cancer metastasis," , Nature reviews. Cancer, vol. 21, no. 7, pp. 446-460, Jul 2021, doi: 10.1038/s41568-021-00353-1.
[21]. P. S. Wang et al., "A novel long non-coding RNA linc-ZNF469-3 promotes lung metastasis through miR-574-5p-ZEB1 axis in triple negative breast cancer," , Oncogene, vol. 37, no. 34, pp. 4662-4678, Aug 2018, doi: 10.1038/s41388-018-0293-1.
[22]. Y. He et al., "LncRNA T376626 is a promising serum biomarker and promotes proliferation, migration, and invasion via binding to LAMC2 in triple-negative breast cancer," , Gene, vol. 860, p. 147227, Apr 15 2023, doi: 10.1016/j.gene.2023.147227.
[23]. X. Liu et al., "Exosomes deliver lncRNA DARS-AS1 siRNA to inhibit chronic unpredictable mild stress-induced TNBC metastasis," , Cancer letters, vol. 543, p. 215781, Sep 1 2022, doi: 10.1016/j.canlet.2022.215781.
[24]. X. Liu et al., "Exosomes deliver lncRNA DARS-AS1 siRNA to inhibit chronic unpredictable mild stress-induced TNBC metastasis," , Cancer letters, vol. 543, p. 215781, Sep 1 2022, doi: 10.1016/j.canlet.2022.215781.
[25]. Y. L. Chen et al., "Novel biomarkers identified in triple-negative breast cancer through RNA-sequencing," , Clinica chimica acta; international journal of clinical chemistry, vol. 531, pp. 302-308, Jun 1 2022, doi: 10.1016/j.cca.2022.04.990.
[26]. J. Kozłowska et al., "Long Intergenic Non-Coding RNAs in HNSCC: From "Junk DNA" to Important Prognostic Factor," , Cancers, vol. 13, no. 12, Jun 12 2021, doi: 10.3390/cancers13122949.
[27]. Y. Li, X. Cao, and H. Li, "Identification and Validation of Novel Long Non-coding RNA Biomarkers for Early Diagnosis of Oral Squamous Cell Carcinoma," , Frontiers in bioengineering and biotechnology, vol. 8, p. 256, 2020, doi: 10.3389/fbioe.2020.00256.
[28]. L. Feng, J. Zhang, M. Sun, F. Qiu, W. Chen, and W. Qiu, "Tumor Suppressor LINC02487 Inhibits Oral Squamous Cell Carcinoma Cell Migration and Invasion Through the USP17-SNAI1 Axis," , Frontiers in oncology, vol. 10, p. 559808, 2020, doi: 10.3389/fonc.2020.559808.
[29]. S. R. Volovat et al., "MiRNA and LncRNA as Potential Biomarkers in Triple-Negative Breast Cancer: A Review," , Frontiers in oncology, vol. 10, p. 526850, 2020, doi: 10.3389/fonc.2020.526850.
[30]. J. S. Mattick et al., "Long non-coding RNAs: definitions, functions, challenges and recommendations," , Nature reviews. Molecular cell biology, vol. 24, no. 6, pp. 430-447, Jun 2023, doi: 10.1038/s41580-022-00566-8.
[31]. X. Yu et al., "Promotion effect of FOXCUT as a microRNA sponge for miR-24-3p on progression in triple-negative breast cancer through the p38 MAPK signaling pathway," , Chinese medical journal, vol. 137, no. 1, pp. 105-114, Jan 5 2024, doi: 10.1097/cm9.0000000000002700.
[32]. R. Sun, S. L. He, H. Y. Liu, X. W. Wang, and S. H. Li, "Tumor Suppressor LINC02487 Inhibits the Progression of Cervical Cancer in Vitro by Regulating the PTEN/Akt/mTOR Pathway," , Discovery medicine, vol. 36, no. 187, pp. 1732-1742, Aug 2024, doi: 10.24976/Discov.Med.202436187.159.
[33]. P. Liu, W. Li, Y. Hu, and Y. Jiang, "Absence of AIF1L contributes to cell migration and a poor prognosis of breast cancer," , OncoTargets and therapy, vol. 11, pp. 5485-5498, 2018, doi: 10.2147/ott.s165874.
[34]. S. Wang et al., "Gene expression in triple-negative breast cancer in relation to survival," , Breast cancer research and treatment, vol. 171, no. 1, pp. 199-207, Aug 2018, doi: 10.1007/s10549-018-4816-9.
[35]. T. Ye et al., "LncRNA MIAT Services as a Noninvasive Biomarker for Diagnosis and Correlated with Immune Infiltrates in Breast Cancer," , International journal of women's health, vol. 13, pp. 991-1004, 2021, doi: 10.2147/ijwh.s312714.









